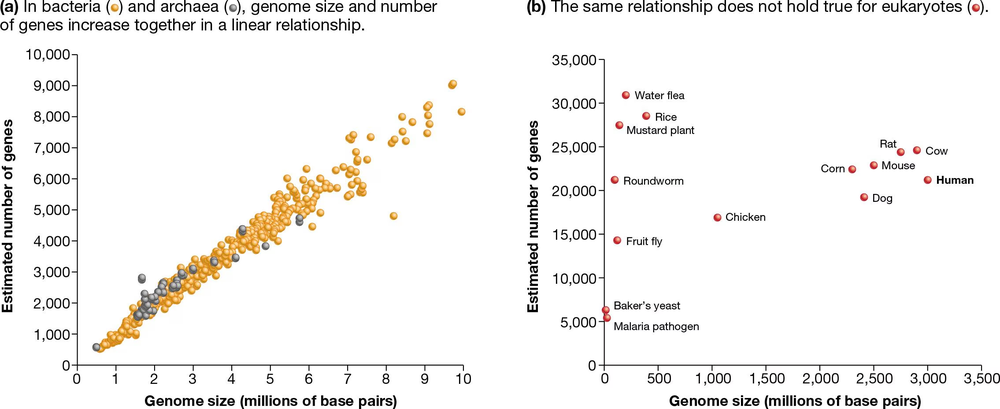
The human genome size is 3 billion base pairs, and the size of the baker's yeast genome, a single-celled organism, is 12 million base pairs. Therefore, the predicted genome size for another single-celled organism, an amoeba,
a. Is about the size of the human genome
b. Is about the size of the yeast genome
c. Is somewhere between the sizes of the yeast and human genomes
d. Cannot be predicted with any certainty