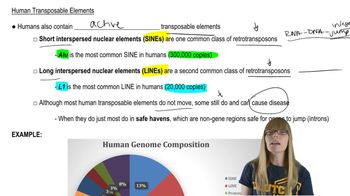
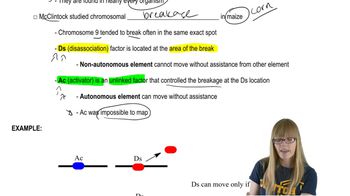
Mammals contain a diploid genome consisting of at least 10⁹ bp. If this amount of DNA is present as chromatin fibers, where each group of 200 bp of DNA is combined with 9 histones into a nucleosome and each group of 6 nucleosomes is combined into a solenoid, achieving a final packing ratio of 50, determine:
(a) the total number of nucleosomes in all fibers,
(b) the total number of histone molecules combined with DNA in the diploid genome, and
(c) the combined length of all fibers.