The pair-rule gene fushi tarazu is expressed in the seven even-numbered parasegments during Drosophila embryogenesis. In contrast, the segment polarity gene engrailed is expressed in the anterior part of each of the 14 parasegments. Since both genes are active at similar times and places during development, it is possible that the expression of one gene is required for the expression of the other. This can be tested by examining the expression of the genes in a mutant background—for example, looking at fushi tarazu expression in an engrailed mutant background, and vice versa. Based on your prediction, can you predict the phenotype of the fushi tarazu and engrailed double mutant?

You are traveling in the Netherlands and overhear a tulip breeder describe a puzzling event. Tulips normally have two outer whorls of brightly colored petal-like organs, a third whorl of stamens, and an inner (fourth) whorl of carpels. However, the breeder found a recessive mutant in his field in which the outer two whorls were green and sepal-like, whereas the third and fourth whorls both contained carpels. What can you speculate about the nature of the gene that was mutated?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Flower Structure and Whorls

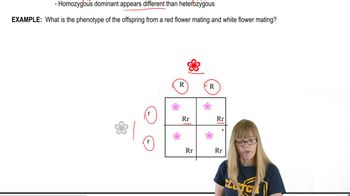
Dominance and Recessiveness in Genetics
Homeotic Genes and Floral Development

In contrast to Drosophila, some insects (e.g., centipedes) have legs on almost every segment posterior to the head. Based on your knowledge of Drosophila, propose a genetic explanation for this phenotype, and describe the expected expression patterns of genes of the Antennapedia and bithorax complexes.
A powerful approach to identifying genes of a developmental pathway is to screen for mutations that suppress or enhance the phenotype of interest. This approach was undertaken to elucidate the genetic pathway controlling C. elegans vulval development. A lin-3 loss-of-function mutant with a vulva-less phenotype was mutagenized. Based on your knowledge of the genetic pathway, what types of mutations will suppress the vulva-less phenotype?
A powerful approach to identifying genes of a developmental pathway is to screen for mutations that suppress or enhance the phenotype of interest. This approach was undertaken to elucidate the genetic pathway controlling C. elegans vulval development. In a complementary experiment, a gain-of-function let-23 mutant with a multi-vulva phenotype was also mutagenized. What types of mutations will suppress the multi-vulva phenotype?
The Hoxd9–13 genes are thought to specify digit identity. What would be the consequence of ectopically expressing Hoxd10 throughout the developing mouse limb bud? What about Hoxd11? What about both Hoxd10 and Hoxd11?
