The human genome is 3×10⁹ bp in length.
How many fragments would be predicted to result from the complete digestion of the human genome with the following enzymes: Sau3A (˘GATC), BamHI (G˘GATCC), EcoRI (G˘AATTC), and NotI (GC˘GGCCGC)?

 Verified step by step guidance
Verified step by step guidance


The human genome is 3×10⁹ bp in length.
How many fragments would be predicted to result from the complete digestion of the human genome with the following enzymes: Sau3A (˘GATC), BamHI (G˘GATCC), EcoRI (G˘AATTC), and NotI (GC˘GGCCGC)?
The human genome is 3×10⁹ bp in length.
How would your initial answer change if you knew that the average GC content of the human genome was 40%?
Ligase catalyzes a reaction between the 5′ phosphate and the 3′ hydroxyl groups at the ends of DNA molecules. The enzyme calf intestinal phosphatase catalyzes the removal of the 5′5′ phosphate from DNA molecules. What would be the consequence of treating a cloning vector, before ligation, with calf intestinal phosphatase?
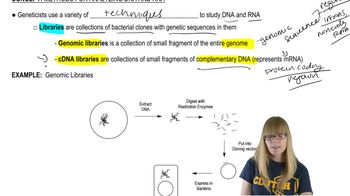
You have constructed four different libraries: a genomic library made from DNA isolated from human brain tissue, a genomic library made from DNA isolated from human muscle tissue, a human brain cDNA library, and a human muscle cDNA library.
Would the sequences contained in each library be expected to overlap completely, partially, or not at all with the sequences present in each of the other libraries?
Using the genomic libraries, you wish to clone the human gene encoding myostatin, which is expressed only in muscle cells.
Assuming the human genome is 3x10⁹ bp and that the average insert size in the genomic libraries is 100 kb, how frequently will a clone representing myostatin be found in the genomic library made from muscle?
Using the genomic libraries, you wish to clone the human gene encoding myostatin, which is expressed only in muscle cells.
How frequently will a clone representing myostatin be found in the genomic library made from brain?