In cattle, an autosomal mutation called Dexter produces calves with short stature and short limbs. Embryos that are homozygous for the Dexter mutation have severely stunted development and either spontaneously abort or are stillborn. What progeny phenotypes do you expect from the cross of two Dexter cows? What are the expected proportions of the expected phenotypes?

The coat color in mink is controlled by two codominant alleles at a single locus. Red coat color is produced by the genotype R₁R₁, silver coat by the genotype R₁R₂, and platinum color by R₂R₂. White spotting of the coat is a recessive trait found with the genotype ss. Solid coat color is found with the S– genotype.
Two crosses are made between mink. Cross 1 is the cross of a solid, silver mink to one that is solid, platinum. Cross 2 is between a spotted, silver mink and one that is solid, silver. The progeny are described in the table below. Use these data to determine the genotypes of the parents in each cross.

 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Codominance
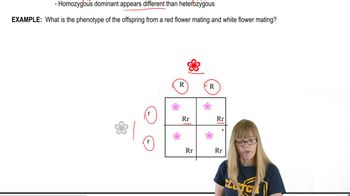
Genotype and Phenotype

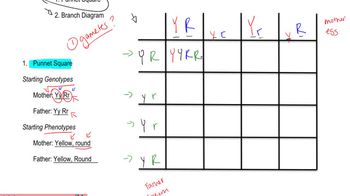
Punnett Square and Genetic Crosses
The coat color in mink is controlled by two codominant alleles at a single locus. Red coat color is produced by the genotype R₁R₁, silver coat by the genotype R₁R₂, and platinum color by R₂R₂. White spotting of the coat is a recessive trait found with the genotype ss. Solid coat color is found with the S– genotype.
What are the expected progeny phenotypes and proportions for the cross SsR₁R₂ x ssR₂R₂?
The coat color in mink is controlled by two codominant alleles at a single locus. Red coat color is produced by the genotype R₁R₁, silver coat by the genotype R₁R₂, and platinum color by R₂R₂. White spotting of the coat is a recessive trait found with the genotype ss. Solid coat color is found with the S– genotype.
If the cross SsR₁R₂ x SsR₁R₁ is made, what are the progeny phenotypes, and in what proportions are they expected to occur?
Strains of petunias come in four pure-breeding colors: white, blue, red, and purple. White petunias are produced when plants synthesize no flower pigment. Blue petunias and red petunias are produced when plants synthesize blue or red pigment only. Purple petunias are produced in plants that synthesize both red and blue pigment (the mixture of red and blue makes purple). Flower-color pigments are synthesized by gene action in two separate pigment-producing biochemical pathways. Pathway I contains gene A that produces an enzyme to catalyze conversion of a colorless pigment designated to blue pigment. In Pathway II, the enzymatic product of gene B converts the colorless pigment designated to red pigment. The two genes assort independently.
What are the possible genotype(s) for pure-breeding red petunias?
Strains of petunias come in four pure-breeding colors: white, blue, red, and purple. White petunias are produced when plants synthesize no flower pigment. Blue petunias and red petunias are produced when plants synthesize blue or red pigment only. Purple petunias are produced in plants that synthesize both red and blue pigment (the mixture of red and blue makes purple). Flower-color pigments are synthesized by gene action in two separate pigment-producing biochemical pathways. Pathway I contains gene A that produces an enzyme to catalyze conversion of a colorless pigment designated to blue pigment. In Pathway II, the enzymatic product of gene B converts the colorless pigment designated to red pigment. The two genes assort independently.
What are the possible genotype(s) for true-breeding blue petunias?
Strains of petunias come in four pure-breeding colors: white, blue, red, and purple. White petunias are produced when plants synthesize no flower pigment. Blue petunias and red petunias are produced when plants synthesize blue or red pigment only. Purple petunias are produced in plants that synthesize both red and blue pigment (the mixture of red and blue makes purple). Flower-color pigments are synthesized by gene action in two separate pigment-producing biochemical pathways. Pathway I contains gene A that produces an enzyme to catalyze conversion of a colorless pigment designated to blue pigment. In Pathway II, the enzymatic product of gene B converts the colorless pigment designated to red pigment. The two genes assort independently.
True-breeding red petunias are crossed to pure-breeding blue petunias, and all the F₁ progeny have purple flowers. If the F₁ are allowed to self-fertilize and produce the F₂, what is the expected phenotypic distribution of the F₂ progeny? Show your work.
