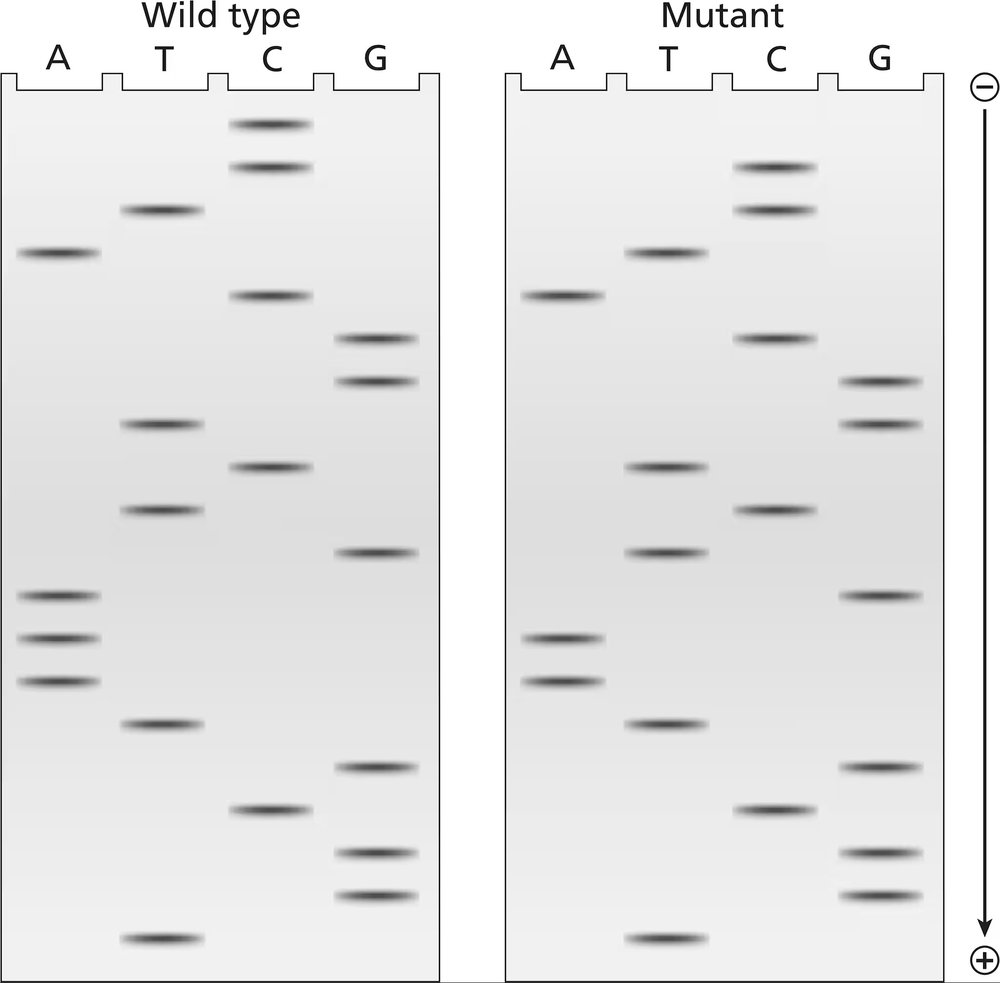
The two gels illustrated contain dideoxynucleotide DNA-sequencing information for a wild-type segment and mutant segment of DNA corresponding to the N-terminal end of a protein. The start codon and the next five codons are sequenced.
Write the DNA sequence of both alleles, including strand polarity.