The two-hybrid method facilitates the discovery of protein–protein interactions. How does this technique work? Can you think of reasons for obtaining a false-positive result, that is, where the proteins encoded by two clones interact in the two-hybrid system but do not interact in the organism in which they naturally occur? Can you think of reasons you might obtain a false-negative result, in which the two proteins interact in vivo but fail to interact in the two-hybrid system?

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 16 - Genomics: Genetics from a Whole-Genome Perspective
Ch. 16 - Genomics: Genetics from a Whole-Genome Perspective Problem 16a
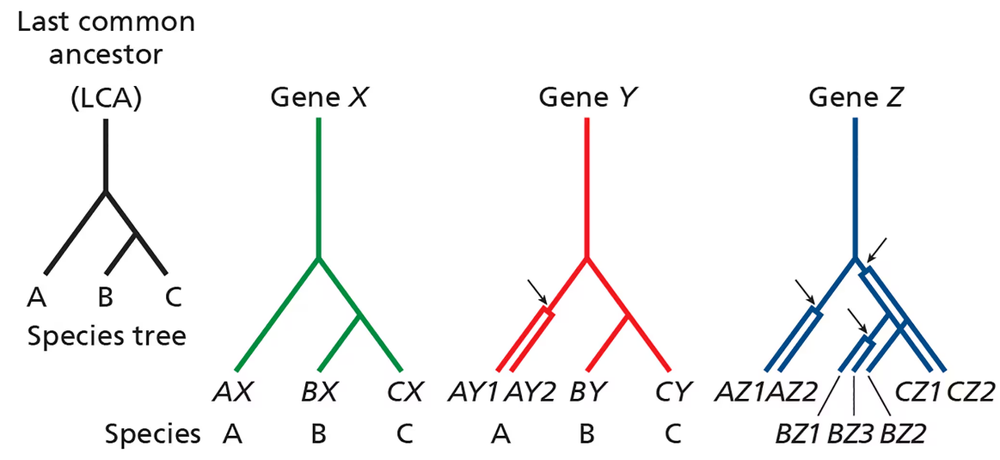
Problem 16aConsider the phylogenetic trees below pertaining to three related species (A, B, and C) that share a common ancestor (last common ancestor, or LCA). The lineage leading to species A diverges before the divergence of species B and C.
For gene X, no gene duplications have occurred in any lineage, and each gene X is derived from the ancestral gene X via speciation events. Are genes AX, BX, and CX orthologous, paralogous, or homologous?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Orthologs

Paralogs

Homologs

Go to http://blast.ncbi.nlm.nih.gov/Blast.cgi and follow the links to nucleotide BLAST. Type in the sequence below; it is broken up into codons to make it easier to copy.
5' ATG TTC GTC AAT CAG CAC CTT TGT GGT TCT CAC CTC GTT GAA GCTTTG TAC CTT GTT TGC GGT GAA CGT GGT TTC TTC TAC ACT CCT AAG ACT TAA 3'
As you will note on the BLAST page, there are several options for tailoring your query to obtain the most relevant information. Some are related to which sequences to search in the database. For example, the search can be limited taxonomically (e.g., restricted to mammals) or by the type of sequences in the database (e.g., cDNA or genomic). For our search, we will use the broadest database, the 'Nucleotide collection (nr/nt).' This is the nonredundant (nr) database of all nucleotide data (nt) in GenBank and can be selected in the 'Database' dialogue box. Other parameters can also be adjusted to make the search more or less sensitive to mismatches or gaps. For our purposes, we will use the default setting, which is automatically presented. Press 'BLAST' to search. What can you say about the DNA sequence?
In the course of the Drosophila melanogaster genome project, the following genomic DNA sequences were obtained. Try to assemble the sequences into a single contig.
5' TTCCAGAACCGGCGAATGAAGCTGAAGAAG 3'
5' GAGCGGCAGATCAAGATCTGGTTCCAGAAC 3'
5' TGATCTGCCGCTCCGTCAGGCATAGCGCGT 3'
5' GGAGAATCGAGATGGCGCACGCGCTATGCC 3'
5' GGAGAATCGAGATGGCGCACGCGCTATGCC 3'
5' CCATCTCGATTCTCCGTCTGCGGGTCAGAT 3'
Go to the URL provided in Problem 14, and using the sequence you have just assembled, perform a blastn search in the 'Nucleotide collection (nr/nt)' database. Does the search produce sequences similar to your assembled sequence, and if so, what are they? Can you tell if your sequence is transcribed, and if it represents protein-coding sequence? Perform a tblastx search, first choosing the 'Nucleotide collection (nr/nt)' database and then limiting the search to human sequences by typing Homo sapiens in the organism box. Are homologous sequences found in the human genome? Annotate the assembled sequence.
Consider the phylogenetic trees below pertaining to three related species (A, B, and C) that share a common ancestor (last common ancestor, or LCA). The lineage leading to species A diverges before the divergence of species B and C.
For gene Y, a gene duplication occurred in the lineage leading to A after it diverged from that, leading to B and C. Are genes AY1 and AY2 orthologous or paralogous? Are genes AY1 and BY orthologous or paralogous? Are genes BY and CY orthologous or paralogous?
Consider the phylogenetic trees below pertaining to three related species (A, B, and C) that share a common ancestor (last common ancestor, or LCA). The lineage leading to species A diverges before the divergence of species B and C.
For gene Z, gene duplications have occurred in all species. Define orthology and paralogy relationships for the different Z genes.
You have isolated a gene that is important for the production of milk and wish to study its regulation. You examine the genomes of human, mouse, dog, chicken, pufferfish, and yeast and note that all genomes except yeast have an orthologous gene.
What does the existence of orthologous genes in chicken and pufferfish tell you about the function of this gene?