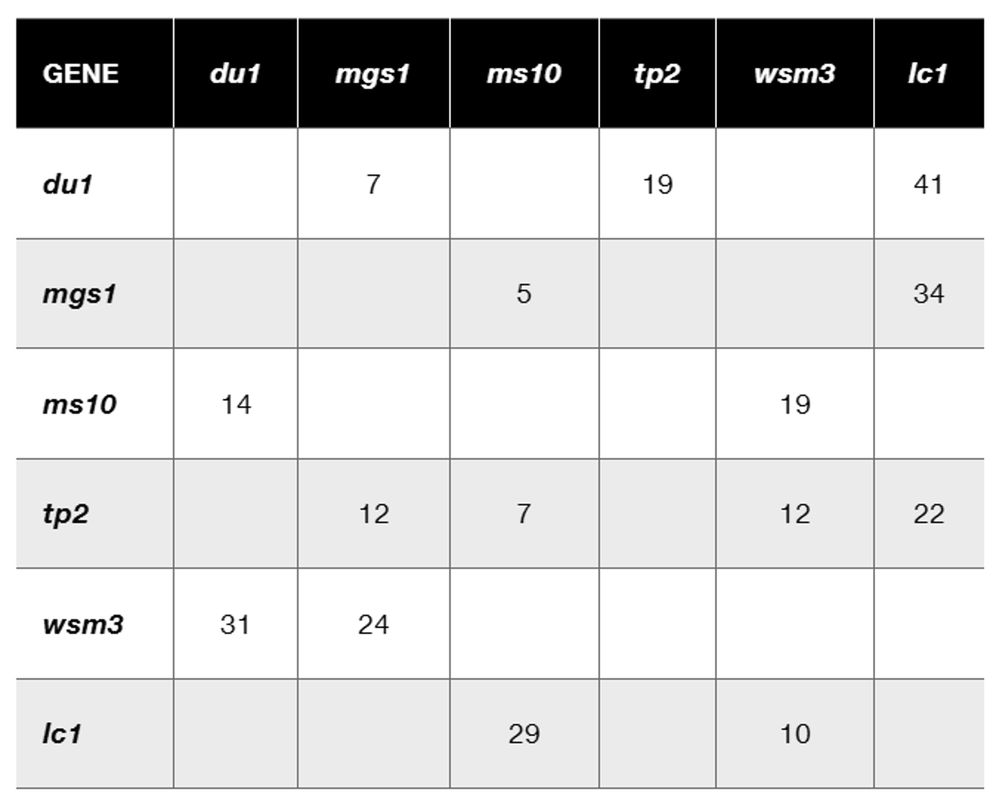
Based on previous family studies, an autosomal recessive disease with alleles A and a is suspected to be linked to an RFLP marker. The RFLP marker has four alleles, R₁, R₂, R₃, and R₄. The accompanying pedigree shows a three-generation family in which the disease is present. The gel shows the RFLP alleles for each family member directly below the pedigree symbol for that person. After determining the genotypes for the RFLP and disease gene for each family member, answer the following questions.
Is there any evidence of recombination in this pedigree? If so, identify the recombinant individuals and illustrate the recombination that has occurred.