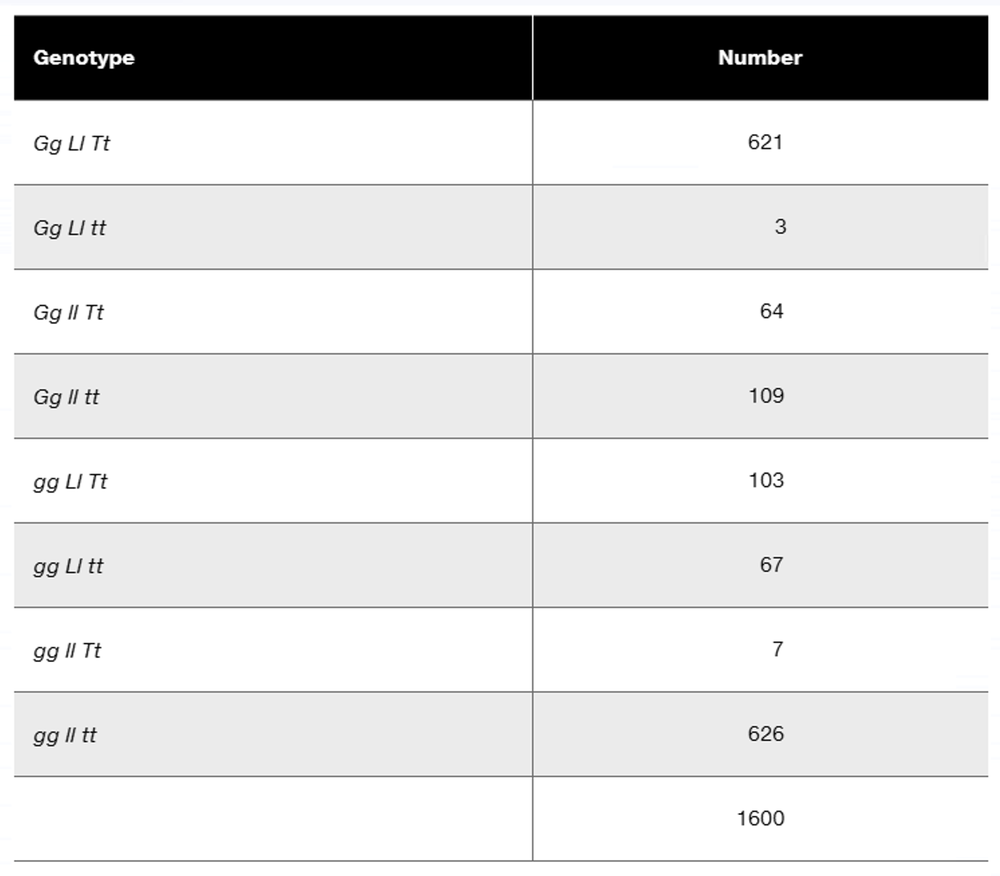
In a diploid plant species, an F₁ with the genotype Gg Ll Tt is test-crossed to a pure-breeding recessive plant with the genotype gg ll tt. The offspring genotypes are as follows:
What is the order of these three linked genes?

 Verified step by step guidance
Verified step by step guidance


In a diploid plant species, an F₁ with the genotype Gg Ll Tt is test-crossed to a pure-breeding recessive plant with the genotype gg ll tt. The offspring genotypes are as follows:
What is the order of these three linked genes?
In a diploid plant species, an F₁ with the genotype Gg Ll Tt is test-crossed to a pure-breeding recessive plant with the genotype gg ll tt. The offspring genotypes are as follows:
Calculate the recombination frequency between each pair of genes.
In a diploid plant species, an F₁ with the genotype Gg Ll Tt is test-crossed to a pure-breeding recessive plant with the genotype gg ll tt. The offspring genotypes are as follows:
Why is the recombination frequency for the outside pair of genes not equal to the sum of recombination frequencies between the adjacent gene pairs?
In a diploid plant species, an F₁ with the genotype Gg Ll Tt is test-crossed to a pure-breeding recessive plant with the genotype gg ll tt. The offspring genotypes are as follows:
Explain the meaning of this I value.
The table given here lists the arrangement of alleles of linked genes in dihybrid organisms, the recombination frequency between the genes, and specific gamete genotypes. Using the information provided, determine the expected frequency of the listed gametes. Assume one map unit equals 1% recombination and, when three genes are involved, interference is zero.
The Rh blood group in humans is determined by a gene on chromosome 1. A dominant allele produces Rh+ blood type, and a recessive allele generates Rh-. Elliptocytosis is an autosomal dominant disorder that produces abnormally shaped red blood cells that have a short life span resulting in hereditary anemia. A large family with elliptocytosis is tested for genetic linkage of Rh blood group and the disease. The lod score data below are obtained for the family.
From these data, can you conclude that Rh and elliptocytosis loci are genetically linked in this family? Why or why not?