A mutant strain of Salmonella bacteria carries a mutation of the rho protein that has full activity at 37°C but is completely inactivated when the mutant strain is grown at 40°C. Speculate about the kind of differences you would expect to see if you compared a broad spectrum of mRNAs from the mutant strain grown at 37°C and the same spectrum of mRNAs from the strain when grown at 40°C.

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 8 - Molecular Biology of Transcription and RNA Processing
Ch. 8 - Molecular Biology of Transcription and RNA Processing Problem 22b
Problem 22bThe human β-globin wild-type allele and a certain mutant allele are identical in sequence except for a single base-pair substitution that changes one nucleotide at the end of intron 2. The wild-type and mutant sequences of the affected portion of pre-mRNA are
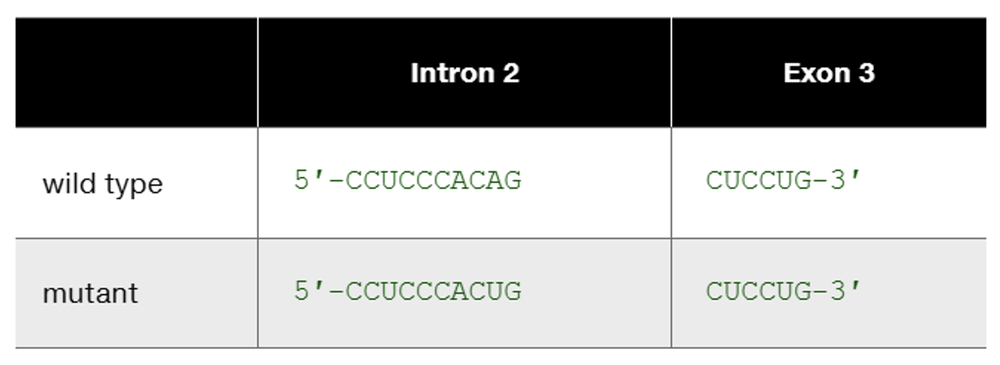
This is one example of how DNA sequence change occurring somewhere other than in an exon can produce mutation. List other kinds of DNA sequence changes occurring outside exons that can produce mutation. In each case, characterize the kind of change you would expect to see in mutant mRNA or mutant protein.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Introns and Exons

Splice Site Mutations

Regulatory Element Mutations
A mutant strain of Salmonella bacteria carries a mutation of the rho protein that has full activity at 37°C but is completely inactivated when the mutant strain is grown at 40°C. Are all mRNAs affected by the rho protein mutation in the same way? Why or why not?
The human β-globin wild-type allele and a certain mutant allele are identical in sequence except for a single base-pair substitution that changes one nucleotide at the end of intron 2. The wild-type and mutant sequences of the affected portion of pre-mRNA are
Speculate about the way in which this base substitution causes mutation of β-globin protein.
Microbiologists describe the processes of transcription and translation as 'coupled' in bacteria. This term indicates that a bacterial mRNA can be undergoing transcription at the same moment it is also undergoing translation.
How is coupling of transcription and translation possible in bacteria?
A full-length eukaryotic gene is inserted into a bacterial chromosome. The gene contains a complete promoter sequence and a functional polyadenylation sequence, and it has wild-type nucleotides throughout the transcribed region. However, the gene fails to produce a functional protein. List at least three possible reasons why this eukaryotic gene is not expressed in bacteria.
A full-length eukaryotic gene is inserted into a bacterial chromosome. The gene contains a complete promoter sequence and a functional polyadenylation sequence, and it has wild-type nucleotides throughout the transcribed region. However, the gene fails to produce a functional protein. What changes would you recommend to permit expression of this eukaryotic gene in a bacterial cell?