Diagram a eukaryotic gene containing three exons and two introns, the pre-mRNA and mature mRNA transcript of the gene, and a partial polypeptide that contains the following sequences and features. Carefully align the nucleic acids, and locate each sequence or feature on the appropriate molecule.
a. The AG and GU dinucleotides corresponding to intron-exon junctions
b. The +1 nucleotide
c. The 5' UTR and the 3' UTR
d. The start codon sequence
e. A stop codon sequence
f. A codon sequence for the amino acids Gly-His-Arg at the end of exon 1 and a codon sequence for the amino acids Leu-Trp-Ala at the beginning of exon 2

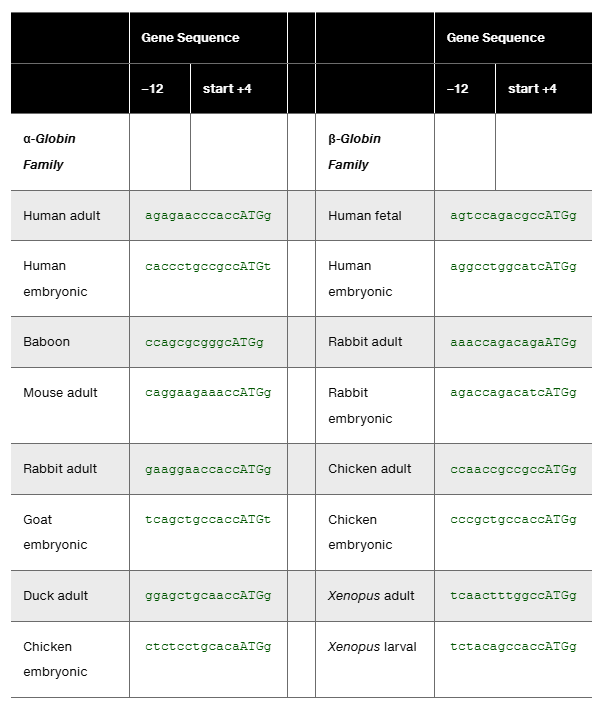
Table D lists α-globin and β-globin gene sequences for the 11 or 12 nucleotides preceding the start codon and the first nucleotide following the start codon (see Problem 34). The data are for 16 vertebrate globin genes reported by Kozak (1987). The sequences are written from -12 to +4 with the start codon sequence in capital letters. Use the data in this table to:
Compare the consensus sequence for these globin genes to the consensus sequence derived from the larger study of 699 vertebrate genes in Problem 34.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Consensus Sequence

Start Codon and Flanking Regions

Comparative Sequence Analysis

Table C contains DNA-sequence information compiled by Marilyn Kozak (1987). The data consist of the percentage of A, C, G, and T at each position among the 12 nucleotides preceding the start codon in 699 genes from various vertebrate species and at the first nucleotide after the start codon. (The start codon occupies positions +1 to +3 and the first nucleotide immediately after the start codon occupies position +4) Use the data to determine the consensus sequence for the 13 nucleotides ( -12 to -1 and +4) surrounding the start codon in vertebrate genes.
Table D lists α-globin and β-globin gene sequences for the 11 or 12 nucleotides preceding the start codon and the first nucleotide following the start codon (see Problem 34). The data are for 16 vertebrate globin genes reported by Kozak (1987). The sequences are written from -12 to +4 with the start codon sequence in capital letters. Use the data in this table to:
Determine the consensus sequence for the 16 selected α-globin and β-globin genes.
The six nucleotides preceding the start codon and the first nucleotide after the start codon in eukaryotes exhibit strong sequence conservation as determined by the percentages of nucleotides in the to positions and the position (see Problem 34). Use the data given in the table for Problem 35 to determine the seven nucleotides that most commonly surround the start in vertebrates.
In terms of the polycistronic composition of mRNAs and the presence or absence of Shine–Dalgarno sequences, compare and contrast bacterial, archaeal, and eukaryotic mRNAs.
Organisms of all three domains of life usually use the mRNA codon AUG as the start codon.
Do organisms of the three domains use the same amino acid as the initial amino acid in translation? Identify similarities and differences.
