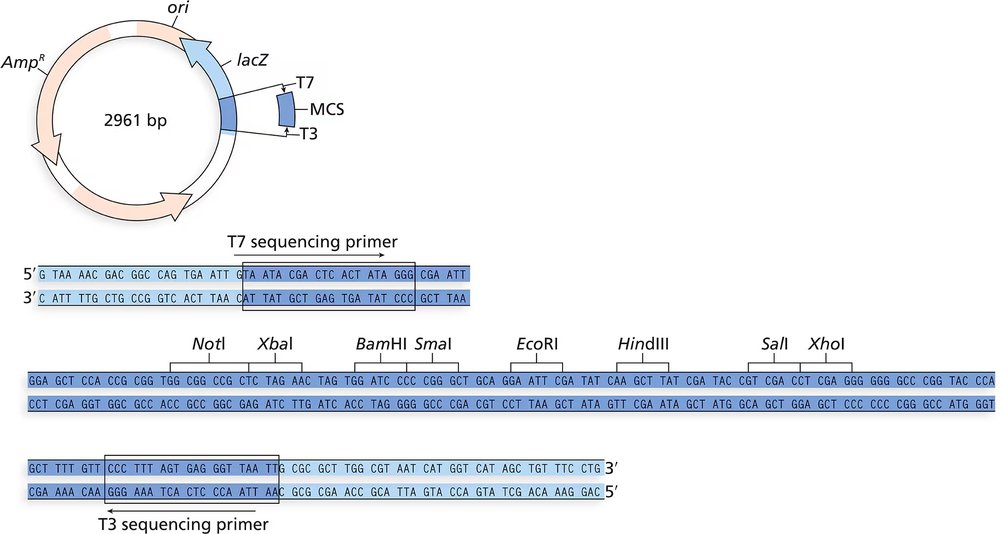
To further analyze the CRABS CLAW gene, you create a map of the genomic clone. The 11-kb EcoRI fragment is ligated into the EcoRI site of the MCS of the vector shown in Problem 18. You digest the double-stranded form of the genome with several restriction enzymes and obtain the following results. Draw, as far as possible, a map of the genomic clone of CRABS CLAW.
What restriction digest would help resolve any ambiguity in the map?