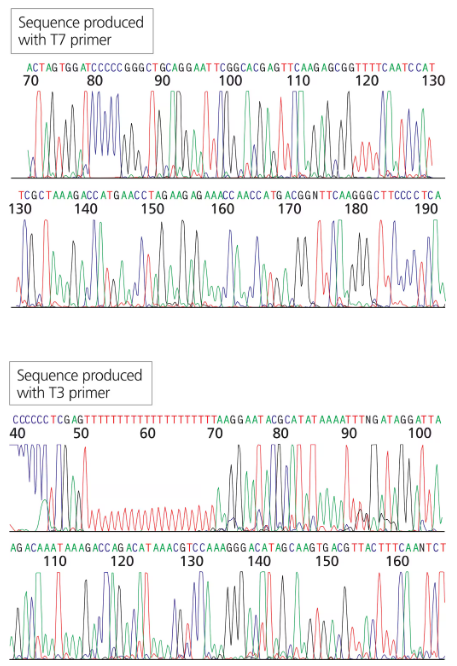
You have isolated a genomic clone with an EcoRI fragment of 11 kb that encompasses the CRABS CLAW gene (see Problem 18). You digest the genomic clone with HindIII and note that the 11-kb EcoRI fragment is split into three fragments of 9 kb, 1.5 kb, and 0.5 kb.
Restriction enzyme sites within a cDNA clone are often also found in the genomic sequence. Can you think of a reason why occasionally this is not the case? What about the converse: Are restriction enzyme sites in a genomic clone always in a cDNA clone of the same gene?