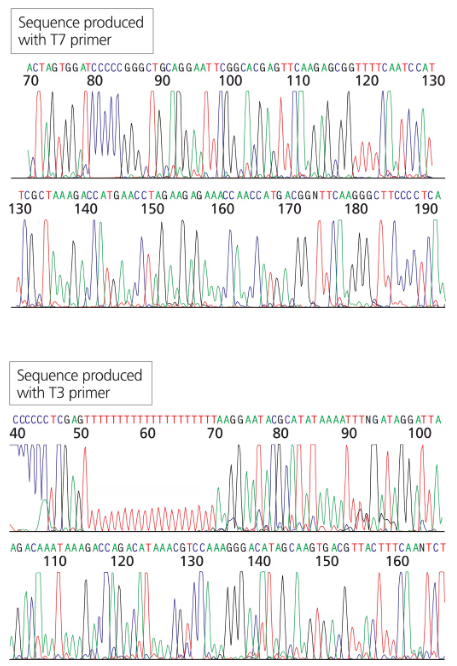
You have isolated a genomic clone with an EcoRI fragment of 11 kb that encompasses the CRABS CLAW gene. You digest the genomic clone with HindIII and note that the 11-kb EcoRI fragment is split into three fragments of 9 kb, 1.5 kb, and 0.5 kb.
Does this tell you anything about where the CRABS CLAW gene is located within the 11-kb genomic clone?