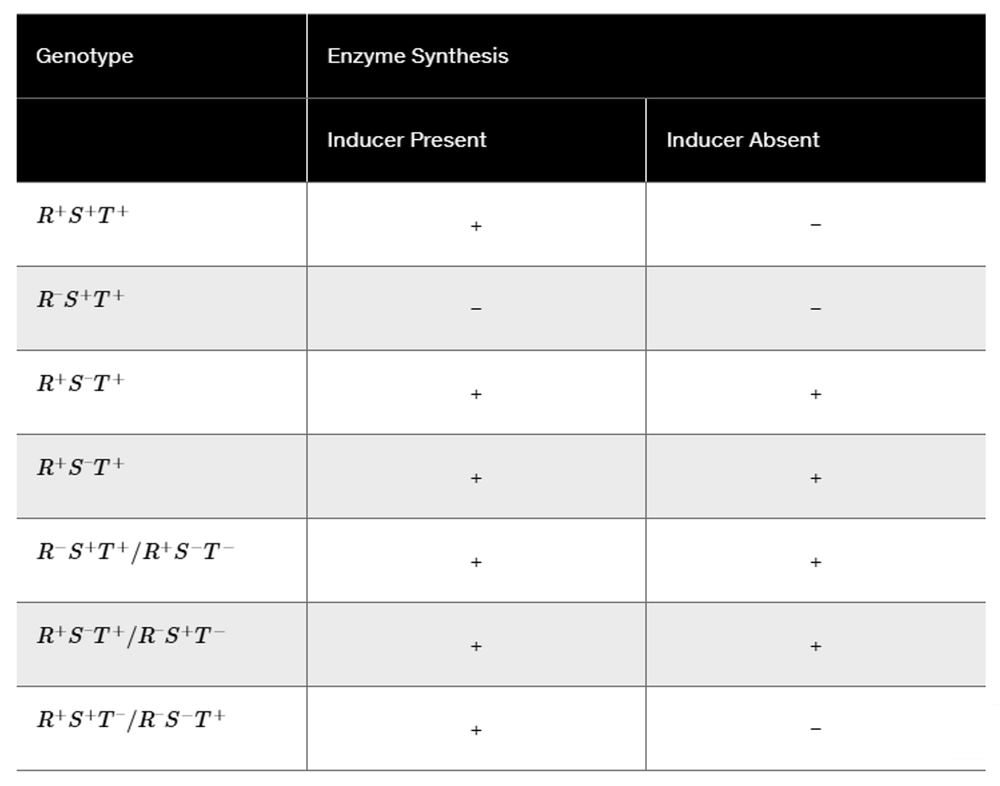
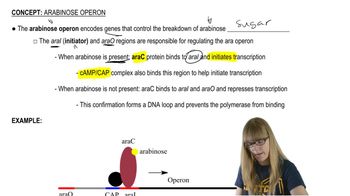
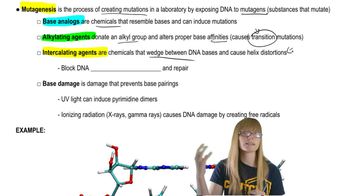
Northern blot analysis is performed on cellular mRNA isolated from E. coli. The probe used in the northern blot analysis hybridizes to a portion of the lacY sequence. Below is an example of the gel from northern blot analysis for a wild-type lac⁺ bacterial strain. In this gel, lane 1 is from bacteria grown in a medium containing only glucose (minimal medium). Lane 2 is from bacteria in a medium containing only lactose. Following the style of this diagram, draw the gel appearance for northern blots of the bacteria listed below. In each case, lane 1 is for mRNA isolated after growth in a glucose-containing (minimal) medium, and lane 2 is for mRNA isolated after growth in a lactose-only medium.
lac⁻ bacteria with the genotype I⁺ P⁺ O⁺ Z⁻ Y⁺ that has a polar mutation affecting the lacZ gene