A bacterial inducible operon, similar to the lac operon, contains three genes—R, T, and S—that are involved in coordinated regulation of transcription. One of these genes is an operator region, one is a regulatory protein, and the third produces a structural enzyme. In the table below, '+' indicates that the structural enzyme is synthesized and '−' indicates that it is not produced. Use the information provided to determine which gene is the operator, which produces the regulatory protein, and which produces the enzyme.

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 12 - Regulation of Gene Expression in Bacteria and Bacteriophage
Ch. 12 - Regulation of Gene Expression in Bacteria and Bacteriophage Problem 37b
Problem 37bThe electrophoresis gel shown in part (a) is from a DNase I footprint analysis of an operon transcription control region. DNA sequence analysis of a 35-bp region is shown in part (b). The control region, labeled with ³²P at one end, is shown in a map in part (c). Separate samples of control-region DNA are exposed to DNase I, and the resulting DNase I–digested DNA is run in separate lanes of the electrophoresis gel. Unprotected DNA is in lane 1, DNA protected by repressor protein is in lane 2, and RNA polymerase–protected DNA is in lane 3. The numbers along the electrophoresis gel correspond to the 35-bp sequence labeled on the map in part (c). Use the information provided to solve the following problems.
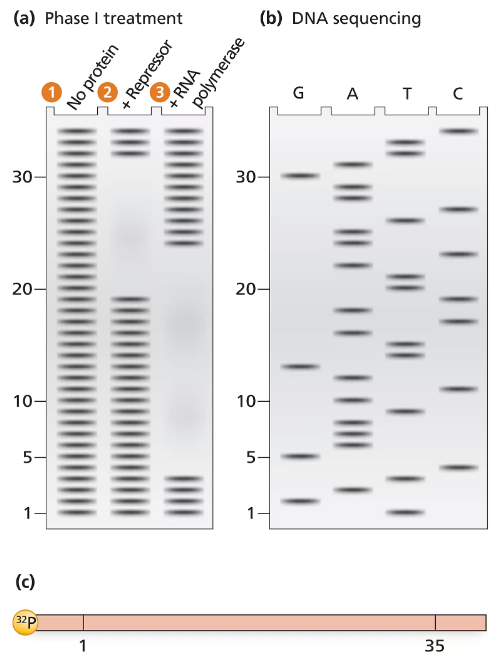
Locate the regions of the sequence protected by repressor protein and by RNA polymerase.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
DNase I Footprint Analysis

Electrophoresis

Transcription Regulation

For the following lac operon partial diploids, determine whether the synthesis of lacZ mRNA is 'constitutive,' 'inducible,' or 'uninducible,' and indicate whether the partial diploid is or (able or not able to utilize lactose).
The electrophoresis gel shown in part (a) is from a DNase I footprint analysis of an operon transcription control region. DNA sequence analysis of a 35-bp region is shown in part (b). The control region, labeled with ³²P at one end, is shown in a map in part (c). Separate samples of control-region DNA are exposed to DNase I, and the resulting DNase I–digested DNA is run in separate lanes of the electrophoresis gel. Unprotected DNA is in lane 1, DNA protected by repressor protein is in lane 2, and RNA polymerase–protected DNA is in lane 3. The numbers along the electrophoresis gel correspond to the 35-bp sequence labeled on the map in part (c). Use the information provided to solve the following problems.
Determine the DNA sequence of the 35-bp region examined.