When the S. cerevisiae genome was sequenced and surveyed for possible genes, only about 40% of those genes had been previously identified in forward genetic screens. This left about 60% of predicted genes with no known function, leading some to dub the genes fun (function unknown) genes. As an approach to understanding the function of a certain fun gene, you wish to create a loss-of-function allele. How will you accomplish this?

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 14 - Analysis of Gene Function via Forward Genetics and Reverse Genetics
Ch. 14 - Analysis of Gene Function via Forward Genetics and Reverse Genetics Problem 16
Problem 16In humans, Duchenne muscular dystrophy is caused by a mutation in the dystrophin gene, which resides on the X chromosome. How would you create a mouse model of this genetic disease?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Duchenne Muscular Dystrophy (DMD)
X-Linked Inheritance
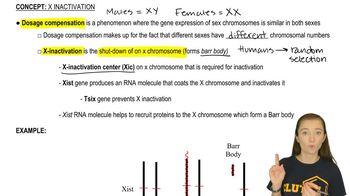
Mouse Models in Genetics

When the S. cerevisiae genome was sequenced and surveyed for possible genes, only about 40% of those genes had been previously identified in forward genetic screens. This left about 60% of predicted genes with no known function, leading some to dub the genes fun (function unknown) genes. You wish to know the physical location of the encoded protein product. How will you obtain such information?
Translational fusions between a protein of interest and a reporter protein are used to determine the subcellular location of proteins in vivo. However, fusion to a reporter protein sometimes renders the protein of interest nonfunctional because the addition of the reporter protein interferes with proper protein folding, enzymatic activity, or protein–protein interactions. You have constructed a fusion between your protein of interest and a reporter gene. How will you show that the fusion protein retains its normal biological function?
How would you perform a genetic screen to identify genes directing Drosophila wing development? Once you have a collection of wing-development mutants, how would you analyze your mutagenesis to learn how many genes are represented and how many alleles of each gene? How would you discover whether the genes act in the same or different pathways, and if in the same pathway, how do you discover the order in which they act? How would you clone the genes?
In enhancer trapping experiments, a minimal promoter and a reporter gene are placed adjacent to the end of a transposon so that genomic enhancers adjacent to the insertion site can act to drive expression of the reporter gene. In a modification of this approach, a series of enhancers and a promoter can be placed at the end of a transposon so that transcription is activated from the transposon into adjacent genomic DNA. What types of mutations do you expect to be induced by such a transposon in a mutagenesis experiment?
We designed a screen to identify conditional mutants of S. cerevisiae in which the secretory system was defective. Suppose we were successful in identifying 12 mutants.
Describe the crosses you would perform to determine the number of different genes represented by the 12 mutations.