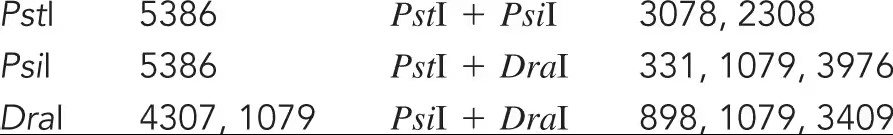The bacteriophage lambda genome can exist in either a linear form or a circular form.
How many fragments will be formed by restriction enzyme digestion with XhoI alone, with XbaI alone, and with both XhoI and XbaI in the linear and circular forms of the lambda genome?