You have sequenced a 100-kb region of the Bacillus anthracis genome (the bacterium that causes anthrax) and a 100-kb region from the Gorilla gorilla genome. What differences and similarities might you expect to see in the annotation of the sequences, for example, in the number of genes, gene structure, regulatory sequences, and repetitive DNA?

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 16 - Genomics: Genetics from a Whole-Genome Perspective
Ch. 16 - Genomics: Genetics from a Whole-Genome Perspective Problem 10
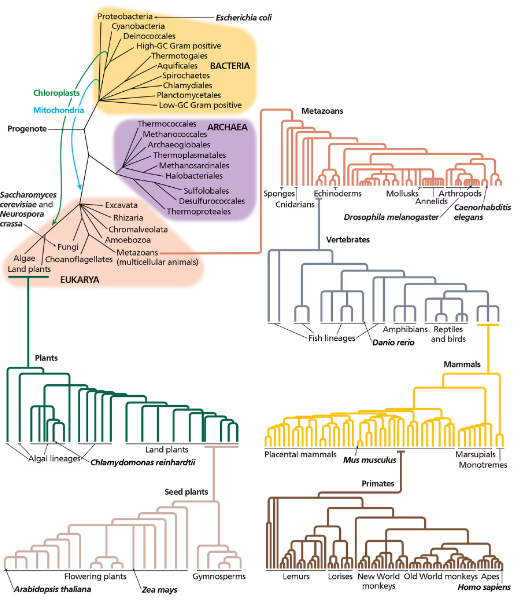
Problem 10Based on the tree of life in the following figure (Figure 16.12), would you expect human proteins to be more similar to fungal proteins or to plant proteins? Would you expect plant proteins to be more similar to fungal proteins or to human proteins?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Phylogenetic Tree
Protein Homology

Common Ancestry

You have just obtained 100 kb of genomic sequence from an as-yet-unsequenced mammalian genome. What are three methods you might use to identify potential genes in the 100 kb? What are the advantages and limitations of each method?
The human genome contains a large number of pseudogenes. How would you distinguish whether a particular sequence encodes a gene or a pseudogene? How do pseudogenes arise?
When comparing genes from two sequenced genomes, how does one determine whether two genes are orthologous? What pitfalls arise when one or both of the genomes are not sequenced?
What is a reference genome? How can it be used to survey genetic variation within a species?
The two-hybrid method facilitates the discovery of protein–protein interactions. How does this technique work? Can you think of reasons for obtaining a false-positive result, that is, where the proteins encoded by two clones interact in the two-hybrid system but do not interact in the organism in which they naturally occur? Can you think of reasons you might obtain a false-negative result, in which the two proteins interact in vivo but fail to interact in the two-hybrid system?