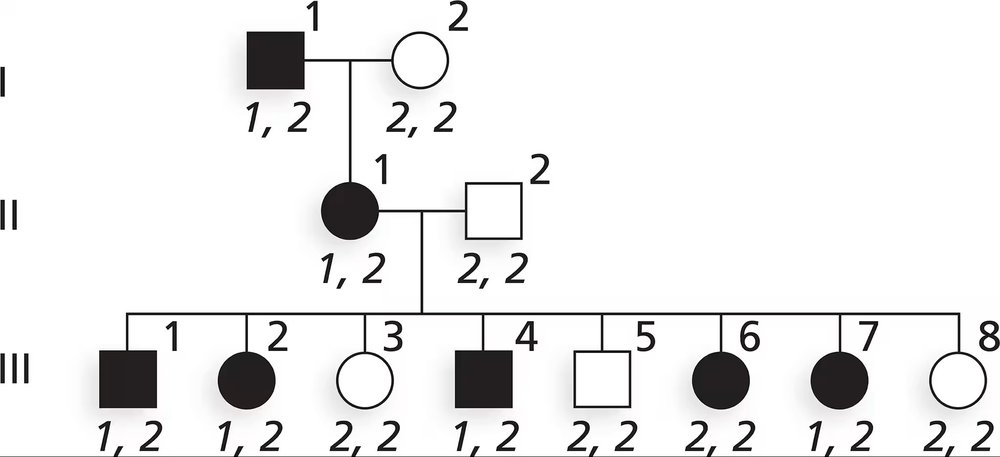
In tomatoes, the allele T for tall plant height is dominant to dwarf allele t, the P allele for smooth skin is dominant to the p allele for peach fuzz skin, and the allele R for round fruit is dominant to the recessive r allele for oblong fruit. The genes controlling these traits are linked on chromosome 1 in the tomato genome, and the genes are arranged in the order and with the recombination frequencies shown.
The F₁ are test-crossed to dwarf, peach fuzz, oblong plants, and 1000 test-cross progeny are produced. What are the phenotypes of test-cross progeny, and what number of progeny is expected in each class?