Telomeres are found at the ends of eukaryotic chromosomes. How does telomerase assemble telomeres?

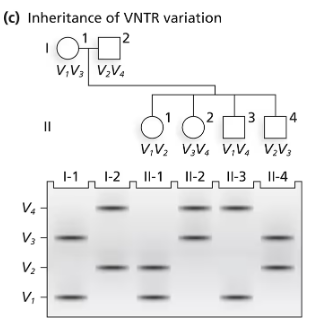
A family consisting of a mother (I-1), a father (I-2), and three children (II-1, II-2, and II-3) are genotyped by PCR for a region of an autosome containing repeats of a 10-bp sequence. The mother carries 16 repeats on one chromosome and 21 on the homologous chromosome. The father carries repeat numbers of 18 and 26.
Following the layout of the following figure, which aligns members of a pedigree with their DNA fragments in a gel, draw a DNA gel containing the PCR fragments generated by amplification of DNA from the parents (I-1 and I-2). Label the size of each fragment.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Polymerase Chain Reaction (PCR)
DNA Fragment Size and Gel Electrophoresis
Genetic Inheritance Patterns

Telomeres are found at the ends of eukaryotic chromosomes. What is the functional role of telomeres?
Telomeres are found at the ends of eukaryotic chromosomes. Why is telomerase usually active in germ-line cells but not in somatic cells?
A family consisting of a mother (I-1), a father (I-2), and three children (II-1, II-2, and II-3) are genotyped by PCR for a region of an autosome containing repeats of a 10-bp sequence. The mother carries 16 repeats on one chromosome and 21 on the homologous chromosome. The father carries repeat numbers of 18 and 26.
Identify all the possible genotypes of children of this couple by specifying PCR fragment lengths in each genotype.
A family consisting of a mother (I-1), a father (I-2), and three children (II-1, II-2, and II-3) are genotyped by PCR for a region of an autosome containing repeats of a 10-bp sequence. The mother carries 16 repeats on one chromosome and 21 on the homologous chromosome. The father carries repeat numbers of 18 and 26.
What genetic term best describes the pattern of inheritance of this DNA marker? Explain your choice.
In a dideoxy DNA sequencing experiment, four separate reactions are carried out to provide the replicated material for DNA sequencing gels. Reaction products are usually run in gel lanes labeled A, T, C, and G.
Identify the nucleotides used in the dideoxy DNA sequencing reaction that produces molecules for the A lane of the sequencing gel.
