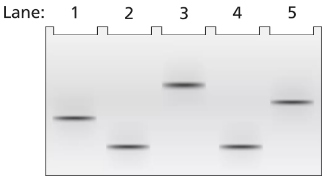
A 2-kb fragment of E. coli DNA contains the complete sequence of a gene for which transcription is terminated by the rho protein. The fragment contains the complete promoter sequence as well as the terminator region of the gene. The cloned fragment is examined by band shift assay. Each lane of a single electrophoresis gel contains the 2-kb cloned fragment under the following conditions:
Lane 1: 2-kb fragment alone
Lane 2: 2-kb fragment plus the core enzyme
Lane 3: 2-kb fragment plus the RNA polymerase holoenzyme
Lane 4: 2-kb fragment plus rho protein
Explain the relative positions of bands in lanes 1 and 4.