A full-length eukaryotic gene is inserted into a bacterial chromosome. The gene contains a complete promoter sequence and a functional polyadenylation sequence, and it has wild-type nucleotides throughout the transcribed region. However, the gene fails to produce a functional protein. What changes would you recommend to permit expression of this eukaryotic gene in a bacterial cell?

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 8 - Molecular Biology of Transcription and RNA Processing
Ch. 8 - Molecular Biology of Transcription and RNA Processing Problem 25c
Problem 25cThe accompanying illustration shows a portion of a gene undergoing transcription. The template and coding strands for the gene are labeled, and a segment of DNA sequence is given.
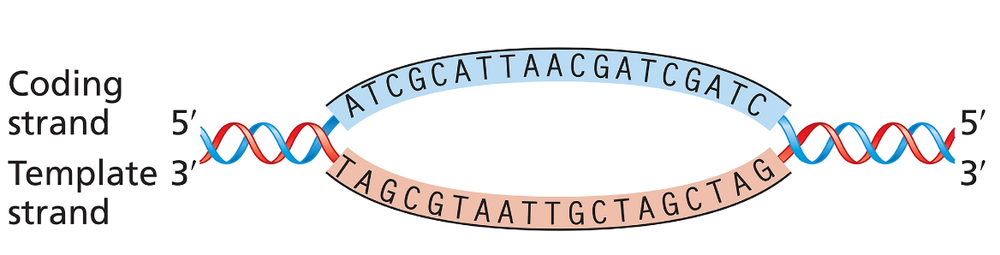
For this gene segment, write the polarity and sequence [TIP 1] of the RNA transcript from the DNA sequence given.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Transcription

Polarity of Nucleic Acids

Base Pairing Rules

The accompanying illustration shows a portion of a gene undergoing transcription. The template and coding strands for the gene are labeled, and a segment of DNA sequence is given.
For this gene segment, superimpose a drawing of RNA polymerase as it nears the end of transcription of the DNA sequence.
The accompanying illustration shows a portion of a gene undergoing transcription. The template and coding strands for the gene are labeled, and a segment of DNA sequence is given.
For this gene segment indicate the direction in which RNA polymerase moves as it transcribes this gene.
The accompanying illustration shows a portion of a gene undergoing transcription. The template and coding strands for the gene are labeled, and a segment of DNA sequence is given.
For this gene segment identify the direction in which the promoter [TIP 2] for this gene is located.
DNA footprint protection is a method that determines whether proteins bind to a specific sample of DNA and thus protect part of the DNA from random enzymatic cleavage by DNase I. A 400-bp segment of cloned DNA is thought to contain a promoter. The cloned DNA is analyzed by DNA footprinting to help determine if it has the capacity to act as a promoter sequence. The accompanying gel has two lanes, each containing the cloned 400-bp DNA fragment treated with DNase I to randomly cleave unprotected DNA. Lane 1 is cloned DNA that was mixed with RNA polymerase II and several TFII transcription factors before exposure to DNase I. Lane 2 contains cloned DNA that was exposed only to DNase I. RNA pol II and TFIIs were not mixed with that DNA before adding DNase I. Explain why this gel provides evidence that the cloned DNA may act as a promoter sequence.
DNA footprint protection is a method that determines whether proteins bind to a specific sample of DNA and thus protect part of the DNA from random enzymatic cleavage by DNase I. A 400-bp segment of cloned DNA is thought to contain a promoter. The cloned DNA is analyzed by DNA footprinting to help determine if it has the capacity to act as a promoter sequence. The accompanying gel has two lanes, each containing the cloned 400-bp DNA fragment treated with DNase I to randomly cleave unprotected DNA. Lane 1 is cloned DNA that was mixed with RNA polymerase II and several TFII transcription factors before exposure to DNase I. Lane 2 contains cloned DNA that was exposed only to DNase I. RNA pol II and TFIIs were not mixed with that DNA before adding DNase I. Approximately what length is the DNA region protected by RNA pol II and TFIIs?