Many antibiotics are effective as drugs to fight off bacterial infections because they inhibit protein synthesis in bacterial cells. Using the information provided in the following table that highlights several antibiotics and their mode of action, discuss which phase of translation is inhibited: initiation, elongation, or termination. What other components of the translational machinery could be targeted to inhibit bacterial protein synthesis?

The flow of genetic information from DNA to protein is mediated by messenger RNA. If you introduce short DNA strands (called antisense oligonucleotides) that are complementary to mRNAs, hydrogen bonding may occur and 'label' the DNA/RNA hybrid for ribonuclease-H degradation of the RNA. One study [Lloyd et al. (2001). Nucl. Acids Res. 29:3664–3673] compared the effect of different-length antisense oligonucleotides upon ribonuclease-H–mediated degradation of tumor necrosis factor (TNFα) mRNA. TNFα exhibits antitumor and pro-inflammatory activities. The following graph indicates the efficacy of various-sized antisense oligonucleotides in causing ribonuclease-H cleavage. What factors other than oligonucleotide length are likely to influence antisense efficacy in vivo?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Antisense Oligonucleotides
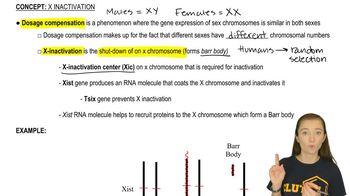
Ribonuclease H (RNase H)

In Vivo Efficacy Factors

The flow of genetic information from DNA to protein is mediated by messenger RNA. If you introduce short DNA strands (called antisense oligonucleotides) that are complementary to mRNAs, hydrogen bonding may occur and 'label' the DNA/RNA hybrid for ribonuclease-H degradation of the RNA. One study [Lloyd et al. (2001). Nucl. Acids Res. 29:3664–3673] compared the effect of different-length antisense oligonucleotides upon ribonuclease-H–mediated degradation of tumor necrosis factor (TNFα) mRNA. TNFα exhibits antitumor and pro-inflammatory activities. The following graph indicates the efficacy of various-sized antisense oligonucleotides in causing ribonuclease-H cleavage. Describe how antisense oligonucleotides interrupt the flow of genetic information in a cell.
The flow of genetic information from DNA to protein is mediated by messenger RNA. If you introduce short DNA strands (called antisense oligonucleotides) that are complementary to mRNAs, hydrogen bonding may occur and 'label' the DNA/RNA hybrid for ribonuclease-H degradation of the RNA. One study [Lloyd et al. (2001). Nucl. Acids Res. 29:3664–3673] compared the effect of different-length antisense oligonucleotides upon ribonuclease-H–mediated degradation of tumor necrosis factor (TNFα) mRNA. TNFα exhibits antitumor and pro-inflammatory activities. The following graph indicates the efficacy of various-sized antisense oligonucleotides in causing ribonuclease-H cleavage. What general conclusion can be drawn from the graph?
Infantile cardiomyopathy is a devastating disorder that is fatal during the first year of life due to defects in the function of heart muscles resulting from mitochondrial dysfunction. A study, performed by Götz et al. [(2011). Am. J. Hum. Genet. 88:635–642), identified two different causative mutations in the gene for mitochondrial alanyl-tRNA synthetase (mtAlaRS). One mutation changes a leucine residue at amino acid position 155 to arginine (p.Leu155Arg). The other mutation changes arginine at position 592 to tryptophan (p.Arg592Trp). The mtAlaRS enzyme has an N-terminal domain (amino acids 36–481) that catalyzes tRNA aminoacylation and an internal editing domain (amino acids 484–782) that catalyzes deacylation in the case that the tRNA is charged with the wrong amino acid.
Consider the position of the disease causing missense mutations in the mtAlaRS gene in the context of the known protein domains of this enzyme. What predictions can you make about how these mutations impair protein synthesis within mitochondria in different ways?
Infantile cardiomyopathy is a devastating disorder that is fatal during the first year of life due to defects in the function of heart muscles resulting from mitochondrial dysfunction. A study, performed by Götz et al. [(2011). Am. J. Hum. Genet. 88:635–642), identified two different causative mutations in the gene for mitochondrial alanyl-tRNA synthetase (mtAlaRS). One mutation changes a leucine residue at amino acid position 155 to arginine (p.Leu155Arg). The other mutation changes arginine at position 592 to tryptophan (p.Arg592Trp). The mtAlaRS enzyme has an N-terminal domain (amino acids 36–481) that catalyzes tRNA aminoacylation and an internal editing domain (amino acids 484–782) that catalyzes deacylation in the case that the tRNA is charged with the wrong amino acid.
Which mutation would you predict has a more severe impairment of translation in mitochondria, and why?
