What is CODIS? Describe the four most important features of genetic markers used in this system.

What is the statistical principle underlying genetic health risk assessment? Why are these assessments not predictive of disease occurrence?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Statistical Probability in Genetic Risk Assessment

Multifactorial Nature of Disease
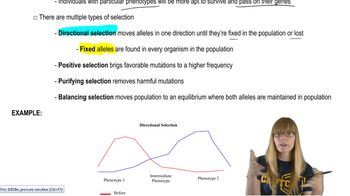
Penetrance and Expressivity

Compare and contrast the terms Paternity Index (PI) and Combined Paternity Index (CPI). How does each contribute to paternity determination?
What is the exclusion principle? How is it used in forensic genetic analysis and in paternity determination?
Explain the meaning of 'identity by descent' in the context of identifying genealogical relationship between individuals. In these analyses, why are segments of chromosomes (haplotypes) rather than individual STRs used to identify genetic relationships?
Figure E.1 illustrates the results of an electrophoretic analysis of 13 CODIS STR markers on a DNA sample and identifies the alleles for each gene. Table E.2 lists the frequencies for alleles of three of the STRs shown in the figure. Use this information to calculate the frequency of the genotype for STR genes FGA, vWA, and D3S1358 given in Figure E.1.
Additional STR allele frequency information can be added to improve the analysis in Problem 8. The frequency of D8S1179₁₂ = 0.12. The frequency of D16S539₁₈ = 0.08 and of D16S539₂₀ = 0.21. Lastly, D18S51₁₉ = 0.13 and D18S51₂₀ = 0.10. Combine the allele frequency information for these three STR genes with the information used in Problem 8 to calculate the frequency of the genotype for six of the STR genes.
