A modification of the two-hybrid system, called the one-hybrid system, is used for identifying proteins that can bind specific DNA sequences. In this method, the DNA sequence to be tested, the bait, is fused to a TATA box to drive expression of a reporter gene. The reporter gene is often chosen to complement a mutant phenotype; for example, a HIS gene may be used in a his⁻ mutant yeast strain. A cDNA library is constructed with the cDNA sequences translationally fused to the GAL4 activation domain and transformed into this yeast strain. Diagram how trans-acting proteins that bind to cis-acting regulatory sequences can be identified using a one-hybrid screen.

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 16 - Genomics: Genetics from a Whole-Genome Perspective
Ch. 16 - Genomics: Genetics from a Whole-Genome Perspective Problem 23
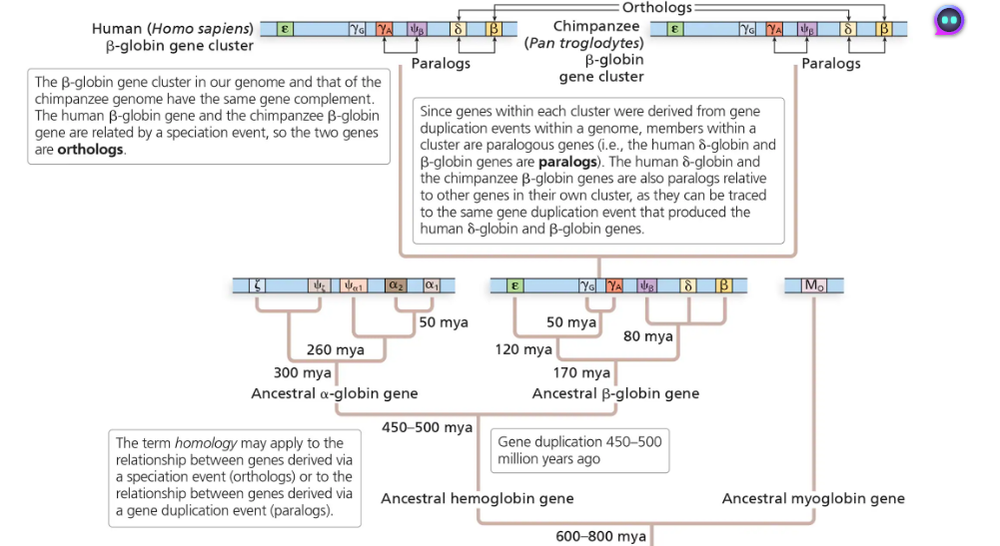
Problem 23In the globin gene family (shown in the below diagram), which pair of genes would exhibit a higher level of sequence similarity, the human δ-globin and human β-globin genes or the human β-globin and chimpanzee β-globin genes? Can you explain your answer in terms of the timing of gene duplications?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Gene Duplication
Sequence Similarity

Phylogenetic Relationships
A substantial fraction of almost every genome sequenced consists of genes that have no known function and that do not have sequence similarity to any genes with known function. Describe two approaches to ascertaining the biological role of these genes in S. cerevisiae.
A substantial fraction of almost every genome sequenced consists of genes that have no known function and that do not have sequence similarity to any genes with known function. How would your approach change if the genes of unknown function were in the human genome?
You are studying similarities and differences in how organisms respond to high salt concentrations and high temperatures. You begin your investigation by using microarrays to compare gene expression patterns of S. cerevisiae in normal growth conditions, in high salt concentrations, and at high temperatures. The results are shown here, with the values of red and green representing the extent of increase and decrease, respectively, of expression for genes a–s in the experimental conditions versus the control (normal growth) conditions. What is the first step you will take to analyze your data?
In conducting the study described in Problem 24, you have noted that a set of S. cerevisiae genes are repressed when yeast are grown under high-salt conditions. How might you approach this question if genome sequences for the related Saccharomyces species S. paradoxus, S. mikatae, and S. bayanus were also available?