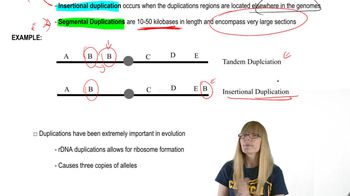
You have isolated a gene that is important for the production of milk and wish to study its regulation. You examine the genomes of human, mouse, dog, chicken, pufferfish, and yeast and note that all genomes except yeast have an orthologous gene.
How would you identify the regulatory elements important for the expression of your isolated gene in mammary glands?