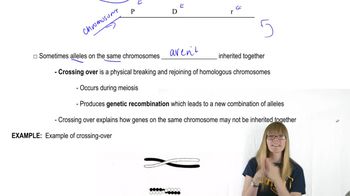
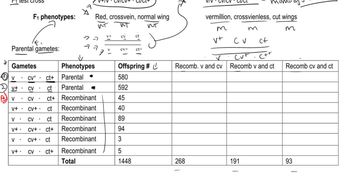
In Drosophila, the map positions of genes are given in map units numbering from one end of a chromosome to the other. The X chromosome of Drosophila is 66 m.u. long. The X-linked gene for body color—with two alleles, y⁺ for gray body and y for yellow body—resides at one end of the chromosome at map position 0.0. A nearby locus for eye color, with alleles w⁺ for red eye and w for white eye, is located at map position 1.5. A third X-linked gene, controlling bristle form, with f⁺ for normal bristles and f for forked bristles, is located at map position 56.7. At each locus the wild-type allele is dominant over the mutant allele.
Explain how each of the predicted progeny classes is produced.