The eukaryotic gene Gen-100 contains four introns labeled A to D. Imagine that Gen-100 has been isolated and its DNA has been denatured and mixed with polyadenylated mRNA from the gene.
Are intron regions single stranded or double stranded? Why?

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 8 - Molecular Biology of Transcription and RNA Processing
Ch. 8 - Molecular Biology of Transcription and RNA Processing Problem 16c
Problem 16c Verified step by step guidance
Verified step by step guidance


The eukaryotic gene Gen-100 contains four introns labeled A to D. Imagine that Gen-100 has been isolated and its DNA has been denatured and mixed with polyadenylated mRNA from the gene.
Are intron regions single stranded or double stranded? Why?
The segment of the bacterial TrpA gene involved in intrinsic termination of transcription is the following:
3'-TGGGTCGGGGCGGATTACTGCCCCGAAAAAAAACTTG-5'
5'-ACCCAGCCCCGCCTAATGACGGGGCTTTTTTTTGAAC-3' Draw the mRNA structure that forms during transcription of this segment of the TrpA gene.
The segment of the bacterial TrpA gene involved in intrinsic termination of transcription is the following:
3'-TGGGTCGGGGCGGATTACTGCCCCGAAAAAAAACTTG-5'
5'-ACCCAGCCCCGCCTAATGACGGGGCTTTTTTTTGAAC-3'
Label the template and coding DNA strands.
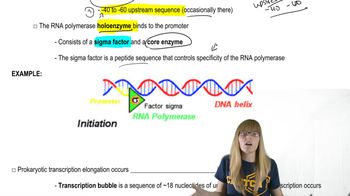
A 2-kb fragment of E. coli DNA contains the complete sequence of a gene for which transcription is terminated by the rho protein. The fragment contains the complete promoter sequence as well as the terminator region of the gene. The cloned fragment is examined by band shift assay. Each lane of a single electrophoresis gel contains the 2-kb cloned fragment under the following conditions:
Lane 1: 2-kb fragment alone
Lane 2: 2-kb fragment plus the core enzyme
Lane 3: 2-kb fragment plus the RNA polymerase holoenzyme
Lane 4: 2-kb fragment plus rho protein
Diagram the relative positions expected for the DNA fragments in this gel electrophoresis analysis.
A 2-kb fragment of E. coli DNA contains the complete sequence of a gene for which transcription is terminated by the rho protein. The fragment contains the complete promoter sequence as well as the terminator region of the gene. The cloned fragment is examined by band shift assay. Each lane of a single electrophoresis gel contains the 2-kb cloned fragment under the following conditions:
Lane 1: 2-kb fragment alone
Lane 2: 2-kb fragment plus the core enzyme
Lane 3: 2-kb fragment plus the RNA polymerase holoenzyme
Lane 4: 2-kb fragment plus rho protein
Explain the relative positions of bands in lanes 1 and 3.
A 2-kb fragment of E. coli DNA contains the complete sequence of a gene for which transcription is terminated by the rho protein. The fragment contains the complete promoter sequence as well as the terminator region of the gene. The cloned fragment is examined by band shift assay. Each lane of a single electrophoresis gel contains the 2-kb cloned fragment under the following conditions:
Lane 1: 2-kb fragment alone
Lane 2: 2-kb fragment plus the core enzyme
Lane 3: 2-kb fragment plus the RNA polymerase holoenzyme
Lane 4: 2-kb fragment plus rho protein
Explain the relative positions of bands in lanes 1 and 4.