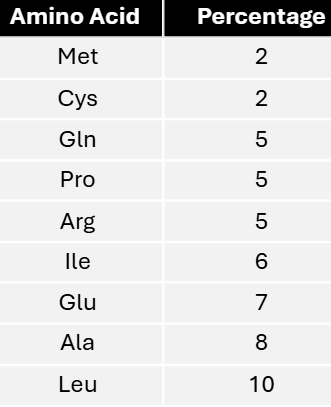
The genetic code is degenerate. Amino acids are encoded by either 1, 2, 3, 4, or 6 triplet codons. An interesting question is whether the number of triplet codes for a given amino acid is in any way correlated with the frequency with which that amino acid appears in proteins. That is, is the genetic code optimized for its intended use? Some approximations of the frequency of appearance of nine amino acids in proteins in E. coli are given in the following:
Determine how many triplets encode each amino acid.