Compare DNA transposons and retrotransposons. What properties do they share?

It is estimated that about 0.2 percent of human mutations are due to TE insertions, and a much higher degree of mutational damage is known to occur in some other organisms. In what way might a TE insertion contribute positively to evolution?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Transposable Elements (TEs)
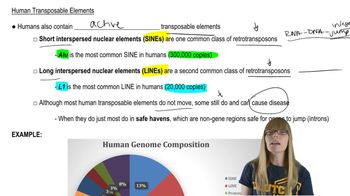
Mutations and Genetic Variation

Positive Evolutionary Impact of TE Insertions

Speculate on how improved living conditions and medical care in the developed nations might affect human mutation rates, both neutral and deleterious.
In maize, a Ds or Ac transposon can alter the function of genes at or near the site of transposon insertion. It is possible for these elements to transpose away from their original insertion site, causing a reversion of the mutant phenotype. In some cases, however, even more severe phenotypes appear, due to events at or near the mutant allele. What might be happening to the transposon or the nearby gene to create more severe mutations?
In a bacterial culture in which all cells are unable to synthesize leucine (leu⁻), a potent mutagen is added, and the cells are allowed to undergo one round of replication. At that point, samples are taken, a series of dilutions are made, and the cells are plated on either minimal medium or minimal medium containing leucine. The first culture condition (minimal medium) allows the growth of only leu⁺ cells, while the second culture condition (minimal medium with leucine added) allows growth of all cells. The results of the experiment are as follows:
What is the rate of mutation at the locus associated with leucine biosynthesis?
Presented here are hypothetical findings from studies of heterokaryons formed from seven human xeroderma pigmentosum cell strains:
These data are measurements of the occurrence or nonoccurrence of unscheduled DNA synthesis in the fused heterokaryon. None of the strains alone shows any unscheduled DNA synthesis. Which strains fall into the same complementation groups? How many different groups are revealed based on these data? What can we conclude about the genetic basis of XP from these data?
