Bacterial strategies to evade natural or human-imposed antibiotics are varied and include membrane-bound efflux pumps that export antibiotics from the cell. A review of efflux pumps [Grkovic, S., et al. (2002)] states that, because energy is required to drive the pumps, activating them in the absence of the antibiotic has a selective disadvantage. The review also states that a given antibiotic may play a role in the regulation of efflux by interacting with either an activator protein or a repressor protein, depending on the system involved. How might such systems be categorized in terms of negative control (inducible or repressible) or positive control (inducible or repressible)?

A marine bacterium is isolated and shown to contain an inducible operon whose genetic products metabolize oil when it is encountered in the environment. Investigation demonstrates that the operon is under positive control and that there is a reg gene whose product interacts with an operator region (o) to regulate the structural genes, designated sg. In an attempt to understand how the operon functions, a constitutive mutant strain and several partial diploid strains were isolated and tested with the results shown in the following table. Host Chromosome F' Factor Phenotype Wild type None Inducible Wild type reg gene from mutant strain Inducible Wild type Operon from mutant strain Constitutive Mutant strain reg gene from wild type Constitutive Draw all possible conclusions about the mutation as well as the nature of regulation of the operon. Is the constitutive mutation in the trans-acting reg element or in the cis-acting o operator element?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Operon Structure and Function

Trans-acting vs. Cis-acting Elements
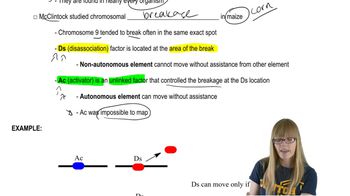
Constitutive Mutations

In a theoretical operon, genes A, B, C, and D represent the repressor gene, the promoter sequence, the operator gene, and the structural gene, but not necessarily in the order named. This operon is concerned with the metabolism of a theoretical molecule (tm). From the data provided in the accompanying table, first decide whether the operon is inducible or repressible. Then assign A, B, C, and D to the four parts of the operon. Explain your rationale. (AE=active enzyme; IE=inactive enzyme; NE=no enzyme.) Genotype tm Present tm Absent A⁺B⁺C⁺D⁺ AE NE A⁻B⁺C⁺D⁺ AE AE A⁺B⁻C⁺D⁺ NE NE A⁺B⁺C⁻D⁺ IE NE A⁺B⁺C⁺D⁻ AE AE A⁻B⁺C⁺D⁺/F'A⁺B⁺C⁺D⁺ AE AE A⁺B⁻C⁺D⁺/F'A⁺B⁺C⁺D⁺ AE NE A⁺B⁺C⁻D⁺/F'A⁺B⁺C⁺D⁺ AE+IE NE A⁺B⁺C⁺D⁻/F'A⁺B⁺C⁺D⁺ AE NE
A bacterial operon is responsible for the production of the biosynthetic enzymes needed to make the hypothetical amino acid tisophane (tis). The operon is regulated by a separate gene, R. The deletion of R causes the loss of enzyme synthesis. In the wild-type condition, when tis is present, no enzymes are made; in the absence of tis, the enzymes are made. Mutations in the operator gene (O⁻) result in repression regardless of the presence of tis. Is the operon under positive or negative control? Propose a model for:
(a) Repression of the genes in the presence of tis in wild-type cells
(b) The mutations.
The SOS repair genes in E. coli are negatively regulated by the lexA gene product, called the LexA repressor. When a cell's DNA sustains extensive damage, the LexA repressor is inactivated by the recA gene product (RecA), and transcription of the SOS genes is increased dramatically. One of the SOS genes is the uvrA gene. You are a student studying the function of the UvrA gene product in DNA repair. You isolate a mutant strain that shows constitutive expression of the UvrA protein. Naming this mutant strain uvrAᶜ, you construct the diagram shown above in the right-hand column showing the lexA and uvrA operons:
Describe two different mutations that would result in a uvrA constitutive phenotype. Indicate the actual genotypes involved.
The SOS repair genes in E. coli are negatively regulated by the lexA gene product, called the LexA repressor. When a cell's DNA sustains extensive damage, the LexA repressor is inactivated by the recA gene product (RecA), and transcription of the SOS genes is increased dramatically. One of the SOS genes is the uvrA gene. You are a student studying the function of the uvrA gene product in DNA repair. You isolate a mutant strain that shows constitutive expression of the UvrA protein. Naming this mutant strain uvrAC, you construct the diagram shown above in the right-hand column showing the lexA and uvrA operons:
Outline a series of genetic experiments that would use partial diploid strains to determine which of the two possible mutations you have isolated.
A fellow student considers the issues in Problem 22 and argues that there is a more straightforward, nongenetic experiment that could differentiate between the two types of mutations. The experiment requires no fancy genetics and would allow you to easily assay the products of the other SOS genes. Propose such an experiment.
