Describe the role of heteroduplex formation during transformation.

 Klug 12th Edition
Klug 12th Edition Ch. 6 - Genetic Analysis and Mapping in Bacteria and Bacteriophages
Ch. 6 - Genetic Analysis and Mapping in Bacteria and Bacteriophages Problem 13
Problem 13Two theoretical genetic strains of a virus (a⁻b⁻c⁻ and a⁺b⁺c⁺) were used to simultaneously infect a culture of host bacteria. Of 10,000 plaques scored, the following genotypes were observed. Determine the genetic map of these three genes on the viral chromosome. Decide whether the interference was positive or negative.

 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Genetic Mapping
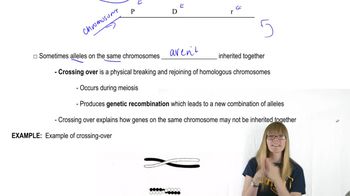
Recombination Frequency

Interference

Explain the observations that led Zinder and Lederberg to conclude that the prototrophs recovered in their transduction experiments were not the result of F⁺ mediated conjugation.
Define plaque, lysogeny, and prophage.
The bacteriophage genome consists of many genes encoding proteins that make up the head, collar, tail, and tail fibers. When these genes are transcribed following phage infection, how are these proteins synthesized, since the phage genome lacks genes essential to ribosome structure?
If a single bacteriophage infects one E. coli cell present on a lawn of bacteria and, upon lysis, yields 200 viable viruses, how many phages will exist in a single plaque if three more lytic cycles occur?
If a single bacteriophage infects one E. coli cell present on a lawn of bacteria and, upon lysis, yields 200 viable viruses, how many phages will exist in a single plaque if three more lytic cycles occur?