Using the adenine–thymine base pair in this DNA sequence
...GCTC...
...CGAG...
Give the sequence after a transversion mutation.

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 11 - Gene Mutation, DNA Repair, and Homologous Recombination
Ch. 11 - Gene Mutation, DNA Repair, and Homologous Recombination Problem 21
Problem 21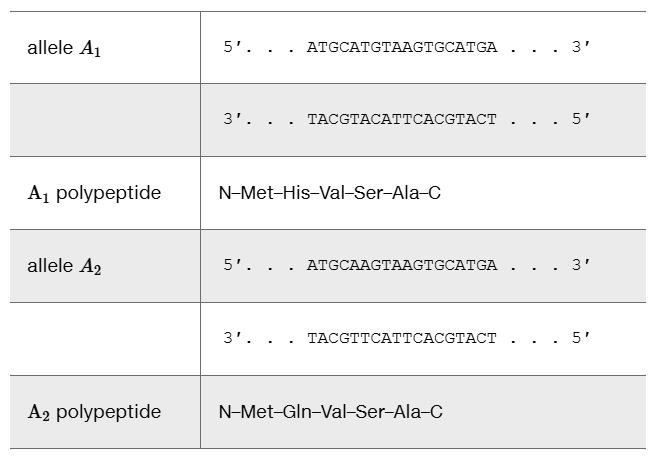
 Verified step by step guidance
Verified step by step guidance


Using the adenine–thymine base pair in this DNA sequence
...GCTC...
...CGAG...
Give the sequence after a transversion mutation.
The partial amino acid sequence of a wild-type protein is
… Arg-Met-Tyr-Thr-Leu-Cys-Ser …
The same portion of the protein from a mutant has the sequence
… Arg-Met-Leu-Tyr-Ala-Leu-Phe …
Identify the type of mutation.
The partial amino acid sequence of a wild-type protein is
… Arg-Met-Tyr-Thr-Leu-Cys-Ser …
The same portion of the protein from a mutant has the sequence
… Arg-Met-Leu-Tyr-Ala-Leu-Phe …
Give the sequence of the wild-type DNA template strand. Use A/G if the nucleotide could be either purine, T/C if it could be either pyrimidine, N if any nucleotide could occur at a site, or the alternative nucleotides if a purine and a pyrimidine are possible.
Many human genes are known to have homologs in the mouse genome. One approach to investigating human hereditary disease is to produce mutations of the mouse homologs of human genes by methods that can precisely target specific nucleotides for mutation.
Numerous studies of mutations of the mouse homologs of human genes have yielded valuable information about how gene mutations influence the human disease process. In general terms, describe how and why creating mutations of the mouse homologs can give information about human hereditary disease processes.
Many human genes are known to have homologs in the mouse genome. One approach to investigating human hereditary disease is to produce mutations of the mouse homologs of human genes by methods that can precisely target specific nucleotides for mutation.
Despite the homologies that exist between human and mouse genes, some attempts to study human hereditary disease processes by inducing mutations in mouse genes indicate there is little to be learned about human disease in this way. In general terms, describe how and why the study of mouse gene mutations might fail to produce useful information about human disease processes.