What is lateral gene transfer? How might it take place between two bacterial cells?

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 6 - Genetic Analysis and Mapping in Bacteria and Bacteriophages
Ch. 6 - Genetic Analysis and Mapping in Bacteria and Bacteriophages Problem 15a
Problem 15aA 2013 CDC report identified the practice of routinely adding antibiotic compounds to animal feed as a major culprit in the rapid increase in the number of antibiotic-resistant strains. Agricultural practice in recent decades has encouraged the addition of antibiotics to animal feed to promote growth rather than to treat disease.
Speculate about the process by which feeding antibiotics to animals such as cattle might lead to an increase in the number of antibiotic-resistant strains of bacteria.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Antibiotic Resistance

Selective Pressure
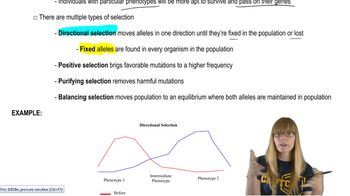
Horizontal Gene Transfer

Lateral gene transfer is thought to have played a major role in the evolution of bacterial genomes. Describe the impact of LGT on bacterial genome evolution.
Seven deletion mutations (1 to 7 in the table below) are tested for their ability to form wild-type recombinants with five point mutations (a to e). The symbol "+" indicates that wild-type recombination occurs, and "-" indicates that wild types are not formed. Use the data to construct a genetic map of the order of point mutations, and indicate the segment deleted by each deletion mutation.
A 2013 CDC report identified the practice of routinely adding antibiotic compounds to animal feed as a major culprit in the rapid increase in the number of antibiotic-resistant strains. Agricultural practice in recent decades has encouraged the addition of antibiotics to animal feed to promote growth rather than to treat disease.
How might the increase in antibiotic-resistant strains of bacteria in cattle be a threat to human health?
Hfr strains that differ in integrated F factor orientation and site of integration are used to construct consolidated bacterial chromosome maps. The data below show the order of gene transfer for five strains.
Hfr Strain Order of Gene Transfer (First → Last)
Hfr A oriT–thr–leu–azi–ton–pro–lac–ade
Hfr B oriT–mtl–xyl–mal–str–his
Hfr C oriT–ile–met–thi–thr–leu–azi–ton
Hfr D oriT–his–trp–gal–ade–lac–pro–ton
Hfr E oriT–thi–met–ile–mtl–xyl–mal–str
Identify the overlaps between Hfr strains. Identify the orientations of integrated F factors relative to one another.
Hfr strains that differ in integrated F factor orientation and site of integration are used to construct consolidated bacterial chromosome maps. The data below show the order of gene transfer for five strains.
Hfr Strain Order of Gene Transfer (First → Last)
Hfr A oriT–thr–leu–azi–ton–pro–lac–ade
Hfr B oriT–mtl–xyl–mal–str–his
Hfr C oriT–ile–met–thi–thr–leu–azi–ton
Hfr D oriT–his–trp–gal–ade–lac–pro–ton
Hfr E oriT–thi–met–ile–mtl–xyl–mal–str
Draw a consolidated map of the bacterial chromosome. (Hint: Begin by placing the insertion site for Hfr A at the 2 o'clock position and arranging the genes thr–leu–azi- . . . in clockwise order.)