Answer these questions concerning promoters.
What consensus sequences are detected in the mammalian β-globin gene promoter?

 Sanders 3rd Edition
Sanders 3rd Edition Ch. 8 - Molecular Biology of Transcription and RNA Processing
Ch. 8 - Molecular Biology of Transcription and RNA Processing Problem 4a
Problem 4a
 Verified step by step guidance
Verified step by step guidance

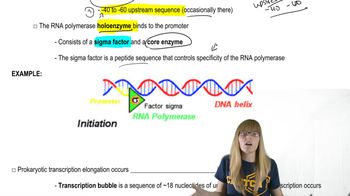

Answer these questions concerning promoters.
What consensus sequences are detected in the mammalian β-globin gene promoter?
Answer these questions concerning promoters.
Eukaryotic promoters are more variable than bacterial promoters. Explain why.
Answer these questions concerning promoters.
What is the meaning of the term alternative promoter? How does the use of alternative promoters affect transcription?
The diagram below shows a DNA duplex. The template strand is identified, as is the location of the nucleotide. consensus sequences.
Assume this region contains a gene transcribed to form mRNA in a eukaryote. Identify the location of the most common promoter.
The diagram below shows a DNA duplex. The template strand is identified, as is the location of the nucleotide.
If this region is a eukaryotic gene transcribed by RNA polymerase III, where are the promoter consensus sequences located?
The following is a portion of an mRNA sequence:
3'-AUCGUCAUGCAGA-5'
During transcription, was the adenine at the left-hand side of the sequence the first or the last nucleotide used to build the portion of mRNA shown? Explain how you know.