An experiment by Khorana and his colleagues translated a synthetic mRNA containing repeats of the trinucelotide UUG.
How does the result of this experiment help confirm the triplet nature of the genetic code?

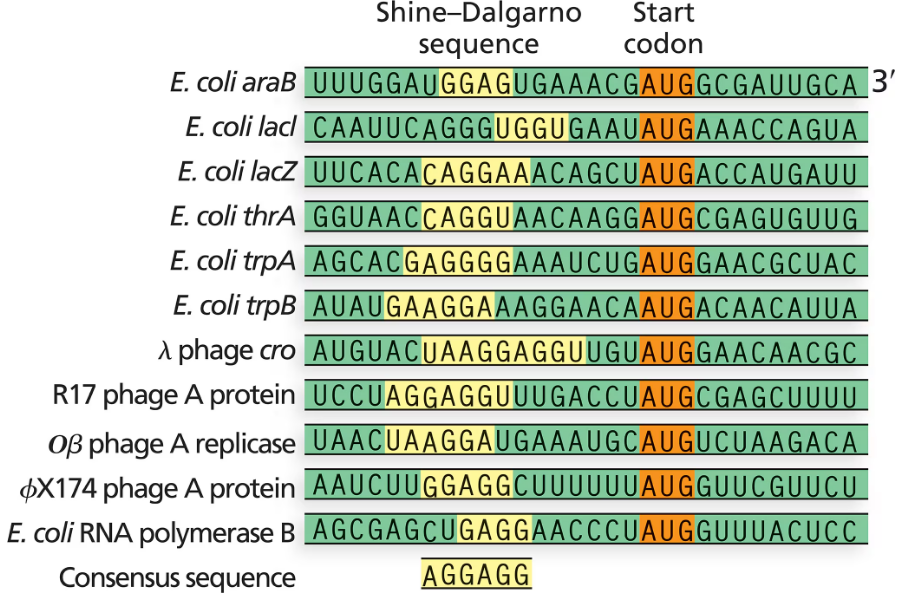
 Verified step by step guidance
Verified step by step guidance



An experiment by Khorana and his colleagues translated a synthetic mRNA containing repeats of the trinucelotide UUG.
How does the result of this experiment help confirm the triplet nature of the genetic code?
The human β-globin polypeptide contains 146 amino acids. How many mRNA nucleotides are required to encode this polypeptide?
The mature mRNA transcribed from the human β-globin gene is considerably longer than the sequence needed to encode the 146–amino acid polypeptide. Give the names of three sequences located on the mature β-globin mRNA but not translated.
A research scientist is interested in producing human insulin in the bacterial species E. coli. Will the genetic code allow the production of human proteins from bacterial cells? Explain why or why not.
Explain why it is not feasible to insert the entire human insulin gene into E. coli and anticipate the production of insulin.
Recombinant human insulin (made by inserting human DNA encoding insulin into E. coli) is one of the most widely used recombinant pharmaceutical products in the world. What segments of the human insulin gene are used to create recombinant bacteria that produce human insulin?