In the context of recombinant DNA technology, of what use is a probe?

What advantages do cDNA libraries provide over genomic DNA libraries? Describe cloning applications where the use of a genomic library is necessary to provide information that a cDNA library cannot.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
cDNA Libraries
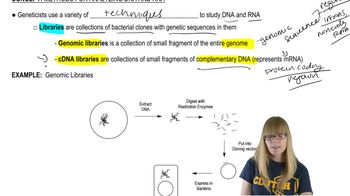
Genomic DNA Libraries

Applications Requiring Genomic Libraries

If you performed a PCR experiment starting with only one copy of double-stranded DNA, approximately how many DNA molecules would be present in the reaction tube after 15 cycles of amplification?
In a control experiment, a plasmid containing a HindIII recognition sequence within a kanamycin resistance gene is cut with HindIII, re-ligated, and used to transform E. coli K12 cells. Kanamycin-resistant colonies are selected, and plasmid DNA from these colonies is subjected to electrophoresis. Most of the colonies contain plasmids that produce single bands that migrate at the same rate as the original intact plasmid. A few colonies, however, produce two bands, one of original size and one that migrates much less far down the gel. Diagram the origin of this slow band as a product of ligation.
You have recovered a cloned DNA segment from a vector and determine that the insert is 1300 bp in length. To characterize this cloned segment, you isolate the insert and decide to construct a restriction map. Using enzyme I and enzyme II, followed by gel electrophoresis, you determine the number and size of the fragments produced by enzymes I and II alone and in combination, as recorded in the following table. Construct a restriction map from these data, showing the positions of the restriction-enzyme cutting sites relative to one another and the distance between them in units of base pairs.
To create a cDNA library, cDNA can be inserted into vectors and cloned. In the analysis of cDNA clones, it is often difficult to find clones that are full length—that is, many clones are shorter than the mature mRNA molecules from which they are derived. Why is this so?
Although the capture and trading of great apes has been banned in 112 countries since 1973, it is estimated that about 1000 chimpanzees are removed annually from Africa and smuggled into Europe, the United States, and Japan. This illegal trade is often disguised by simulating births in captivity. Until recently, genetic identity tests to uncover these illegal activities were not used because of the lack of highly polymorphic markers (markers that vary from one individual to the next) and the difficulties of obtaining chimpanzee blood samples. A study was reported in which DNA samples were extracted from freshly plucked chimpanzee hair roots and used as templates for PCR. The primers used in these studies flank highly polymorphic sites in human DNA that result from variable numbers of tandem nucleotide repeats. Several offspring and their putative parents were tested to determine whether the offspring were 'legitimate' or the product of illegal trading. The data are shown in the following Southern blot.
Examine the data carefully and choose the best conclusion.
a. None of the offspring is legitimate.
b. Offspring B and C are not the products of these parents and were probably purchased on the illegal market. The data are consistent with offspring A being legitimate.
c. Offspring A and B are products of the parents shown, but C is not and was therefore probably purchased on the illegal market.
d. There are not enough data to draw any conclusions. Additional polymorphic sites should be examined.
e. No conclusion can be drawn because 'human' primers were used.
