Explain differences between whole-genome sequencing (WGS) and whole-exome sequencing (WES), and describe advantages and disadvantages of each approach for identifying disease-causing mutations in a genome. Which approach was used for the Human Genome Project?

Metagenomics studies generate very large amounts of sequence data. Provide examples of genetic insight that can be learned from metagenomics.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Metagenomics and Environmental DNA Sequencing

Microbial Diversity and Community Structure

Functional Gene Discovery and Metabolic Potential
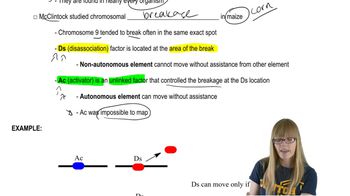
Describe the significance of the Genome 10K project.
It can be said that modern biology is experiencing an 'omics' revolution. What does this mean? Explain your answer.
What are DNA microarrays? How are they used?
Annotation of the human genome sequence reveals a discrepancy between the number of protein-coding genes and the number of predicted proteins actually expressed by the genome. Proteomic analysis indicates that human cells are capable of synthesizing more than 100,000 different proteins and perhaps three times this number. What is the discrepancy, and how can it be reconciled?
An interactive Web site for the Human Proteome Map (HPM) is available at http://www.humanproteomemap.org. Visit this site, and then answer the question.
How many proteins were identified in this project?
