Textbook Question
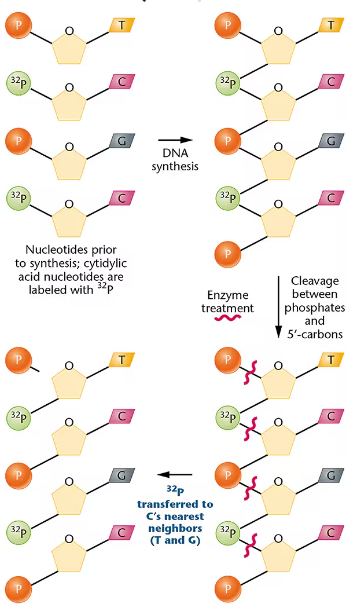
Consider the drawing of a dinucleotide below.
Is it DNA or RNA?
657
views


 Verified step by step guidance
Verified step by step guidance



Consider the drawing of a dinucleotide below.
Is it DNA or RNA?
Consider the drawing of a dinucleotide below.
Is the arrow closest to the 5' or the 3' end?
Consider the drawing of a dinucleotide below.
Suppose that the molecule was cleaved with the enzyme spleen phosphodiesterase, which breaks the covalent bond connecting the phosphate to C-5'. After cleavage, to which nucleoside is the phosphate now attached (A or T)?