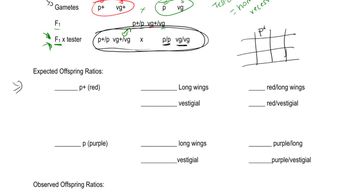
Three independently assorting genes (A, B, and C) are known to control the following biochemical pathway that provides the basis for flower color in a hypothetical plant:
Three homozygous recessive mutations are also known, each of which interrupts a different one of these steps. Determine the phenotypic results in the F1 and F2 generations resulting from the P1 crosses of true-breeding plants listed here:
speckled (AABBCC) × yellow (AAbbCC)