Traditional Sanger sequencing has largely been replaced in recent years by next-generation and third-generation sequencing approaches. Describe advantages of these sequencing methods over first-generation Sanger sequencing.
Ch. 20 - Recombinant DNA Technology

Chapter 20, Problem 24
One complication of making a transgenic animal is that the transgene may integrate at random into the coding region, or the regulatory region, of an endogenous gene. What might be the consequences of such random integrations? How might this complicate genetic analysis of the transgene?
 Verified step by step guidance
Verified step by step guidance1
Understand that when a transgene integrates randomly into the genome, it can insert into different regions such as the coding region (exons) or regulatory region (promoters, enhancers) of an endogenous gene.
If the transgene inserts into the coding region of a gene, it may disrupt the normal function of that gene, potentially causing a loss-of-function mutation or producing a truncated or nonfunctional protein.
If the transgene inserts into the regulatory region of a gene, it may alter the gene's expression pattern, either increasing, decreasing, or misregulating the gene's activity, which can affect the phenotype.
These disruptions can cause unintended phenotypic effects unrelated to the transgene's intended function, making it difficult to distinguish whether observed traits are due to the transgene itself or due to disruption of endogenous genes.
This complicates genetic analysis because multiple independent transgenic lines may show different phenotypes depending on the integration site, requiring careful controls and possibly molecular characterization (e.g., mapping the insertion site) to interpret results accurately.

Verified video answer for a similar problem:
This video solution was recommended by our tutors as helpful for the problem above.
Video duration:
1mWas this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Transgene Integration Sites
Transgenes can insert randomly into the genome, either within coding regions (exons) or regulatory regions (promoters/enhancers) of endogenous genes. This random insertion can disrupt normal gene function or alter gene expression, leading to unintended phenotypic effects.
Recommended video:
Guided course
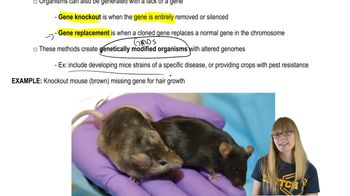
Transgenic Organisms and Gene Therapy
Gene Disruption and Mutagenesis
Insertion of a transgene into an endogenous gene can cause loss-of-function mutations by interrupting coding sequences or regulatory elements. This disruption may produce mutant phenotypes unrelated to the transgene’s intended function, complicating interpretation of experimental results.
Recommended video:
Guided course

Mapping Genes
Complications in Genetic Analysis
Random integration can cause variable expression levels and unpredictable phenotypes due to position effects or gene disruption. This variability makes it difficult to attribute observed traits solely to the transgene, requiring careful controls and multiple independent lines for accurate genetic analysis.
Recommended video:
Guided course

Chi Square Analysis
Related Practice
Textbook Question
520
views
Textbook Question
How is fluorescent in situ hybridization (FISH) used to produce a spectral karyotype?
823
views
Textbook Question
What is the difference between a knockout animal and a transgenic animal?
848
views
Textbook Question
When disrupting a mouse gene by knockout, why is it desirable to breed mice until offspring homozygous (−/−) for the knockout target gene are obtained?
476
views
Textbook Question
What techniques can scientists use to determine if a particular transgene has been integrated into the genome of an organism?
591
views
Textbook Question
Gene targeting and gene editing are both techniques for removing or modifying a particular gene, each of which can produce the same ultimate goal. What is the main technical difference in how DNA is modified that differs between these approaches?
731
views
