What techniques can scientists use to determine if a particular transgene has been integrated into the genome of an organism?

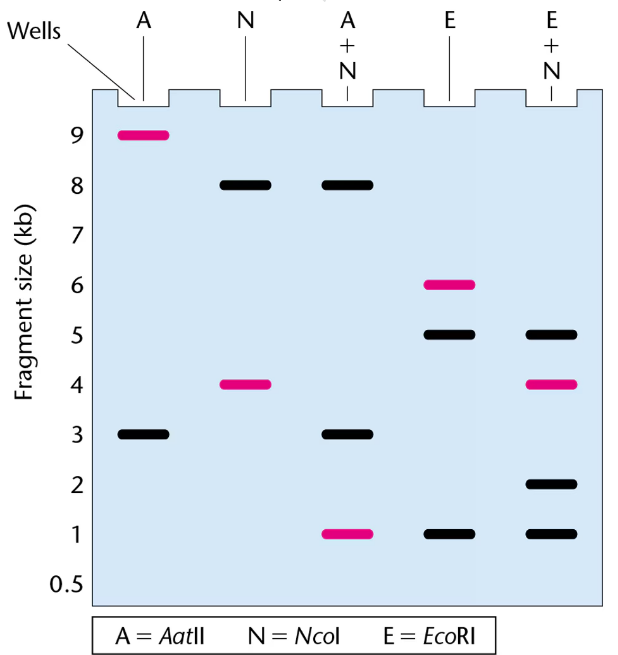
The gel presented here shows the pattern of bands of fragments produced with several restriction enzymes. The enzymes used are identified above the lanes of the gel, and six possible restriction maps are shown in the column to the right.
One of the six restriction maps shown is consistent with the pattern of bands shown in the gel.
From your analysis of the pattern of bands on the gel, select the correct restriction map and explain your reasoning.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Restriction Enzymes and DNA Fragmentation

Gel Electrophoresis and Band Patterns

Restriction Mapping

Gene targeting and gene editing are both techniques for removing or modifying a particular gene, each of which can produce the same ultimate goal. What is the main technical difference in how DNA is modified that differs between these approaches?
The CRISPR-Cas system has great potential but also raises many ethical issues about its potential applications because, theoretically, it can be used to edit any gene in the genome. What do you think are some of the concerns about the use of CRISPR-Cas on humans? Should CRISPR-Cas applications be limited for use on only certain human genes but not others? Explain your answers.
The gel presented here shows the pattern of bands of fragments produced with several restriction enzymes. The enzymes used are identified above the lanes of the gel, and six possible restriction maps are shown in the column to the right.
One of the six restriction maps shown is consistent with the pattern of bands shown in the gel.
The highlighted bands (magenta) in the gel were hybridized with a probe for the gene pep during a Southern blot. Where in the gel is the pep gene located?
A widely used method for calculating the annealing temperature for a primer used in PCR is 5 degrees below the melting temperature, Tₘ(°C), which is computed by the equation 81.5+0.41×(%GC)−(675/N), where %GC is the percentage of GC nucleotides in the oligonucleotide and N is the length of the oligonucleotide. Notice from the formula that both the GC content and the length of the oligonucleotide are variables. Assuming you have the following oligonucleotide as a primer,
5′-TTGAAAATATTTCCCATTGCC-3′
Compute the annealing temperature for PCR. What is the relationship between %GC and? Why? (Note: In reality, this computation provides only a starting point for empirical determination of the most useful annealing temperature.)
Most of the techniques (blotting, cloning, PCR, etc.) are dependent on hybridization (annealing) between different populations of nucleic acids. The length of the strands, temperature, and percentage of GC nucleotides weigh considerably on hybridization. Two other components commonly used in hybridization protocols are monovalent ions and formamide. A formula that takes monovalent Na⁺ ions (M[Na⁺]) and formamide concentrations into consideration to compute a Tₘ (temperature of melting) is as follows:
Tₘ=81.5+16.6(log M[Na+])+0.41(%GC)−0.72(%formamide)
For the following concentrations of Na⁺ and formamide, calculate the Tₘ. Assume 45% GC content.
[Na⁺] % Formamide
0.825 20
0.825 40
0.165 20
0.165 40
