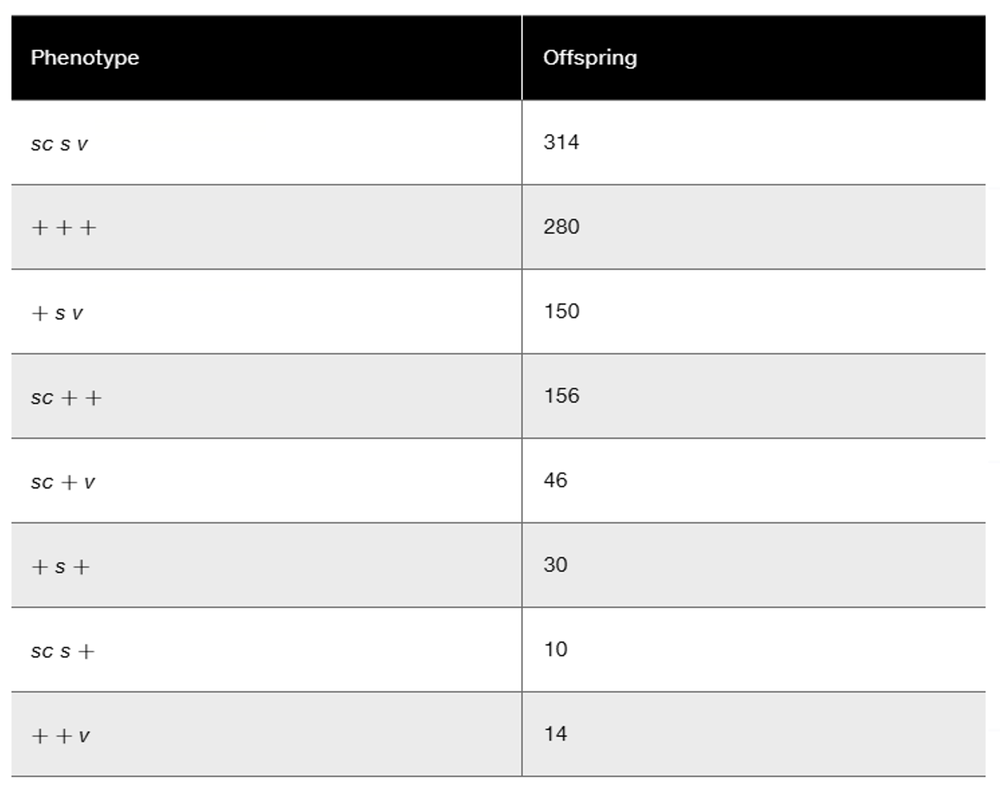
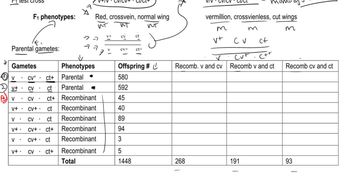
In a series of two-point mapping crosses involving five genes located on chromosome II in Drosophila, the following recombinant (single-crossover) frequencies were observed:
In another set of experiments, a sixth gene, d, was tested against b and pr:
Predict the results of two-point mapping between d and c, d and vg, and d and adp.