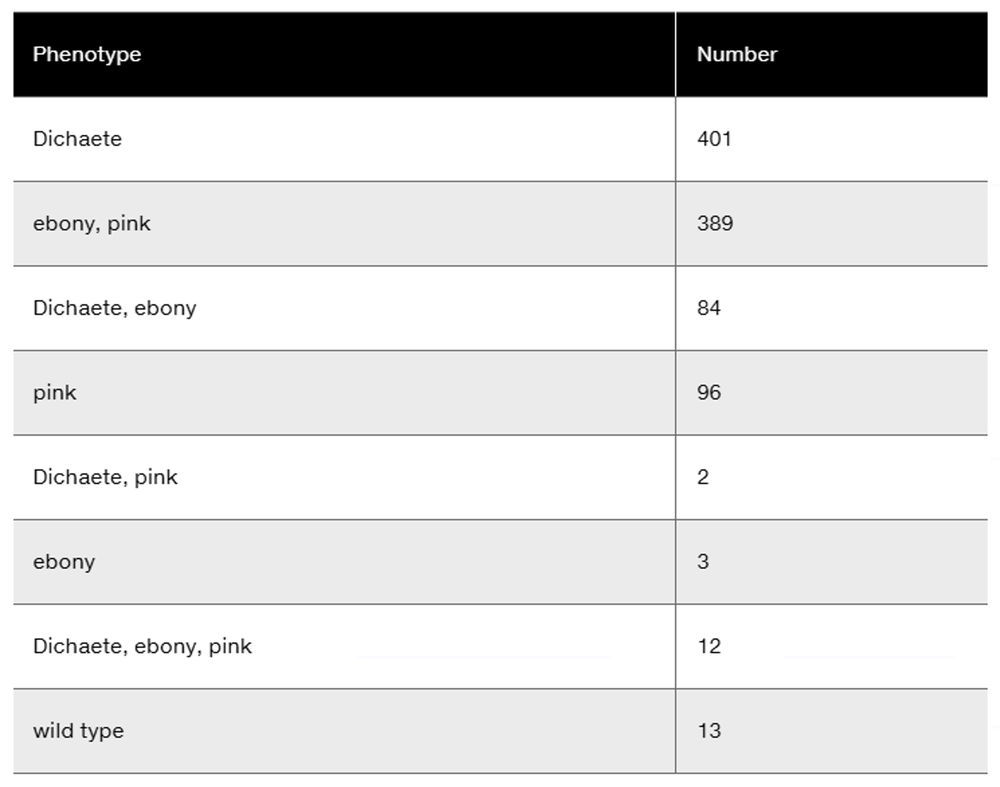
Another cross in Drosophila involved the recessive, X-linked genes yellow (y), white (w), and cut (ct). A yellow-bodied, white-eyed female with normal wings was crossed to a male whose eyes and body were normal but whose wings were cut. The F₁ females were wild type for all three traits, while the F₁ males expressed the yellow-body and white-eye traits. The cross was carried to an F₂ progeny, and only male offspring were tallied. On the basis of the data shown here, a genetic map was constructed.
Were any double-crossover offspring expected?