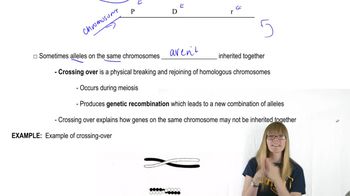
In Drosophila, Dichaete (D) is a mutation on chromosome III with a dominant effect on wing shape. It is lethal when homozygous. The genes ebony body (e) and pink eye (p) are recessive mutations on chromosome III. Flies from a Dichaete stock were crossed to homozygous ebony, pink flies, and the F1 progeny, with a Dichaete phenotype, were backcrossed to the ebony, pink homozygotes. Using the results of this backcross shown in the table,
What is the sequence and interlocus distance between these three genes?