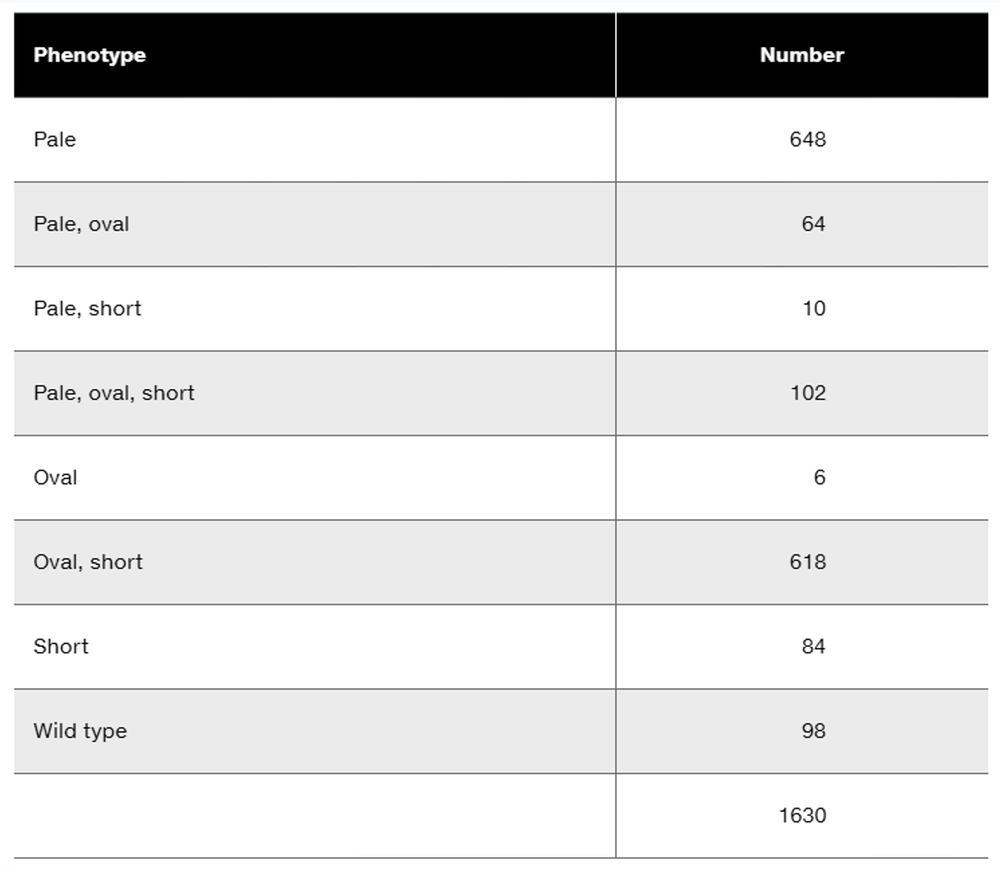
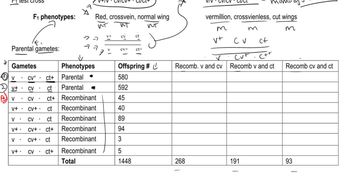
Gene R and gene T are genetically linked. Answer the following questions concerning a dihybrid organism with the genotype Rt/rT:
Can you make a general statement about how the occurrence of two crossover events between a given pair of linked genes affects the estimate of recombination frequency?