The interphase nucleus is a highly structured organelle with chromosome territories, interchromatin compartments, and transcription factories. In cultured human cells, researchers have identified approximately 8000 transcription factories per cell, each containing an average of eight tightly associated RNAP II molecules actively transcribing RNA. If each RNAP II molecule is transcribing a different gene, how might such a transcription factory appear? Provide a simple diagram that shows eight different genes being transcribed in a transcription factory and include the promoters, structural genes, and nascent transcripts in your presentation.

Regulation of the lac operon in E. coli and regulation of the GAL system in yeast are analogous in that they both serve to adapt cells to growth on different carbon sources. However, the transcriptional changes are accomplished very differently. Consider the conceptual similarities and differences as you address the following.
Compare and contrast the roles of the lac operon inducer in bacteria and Gal3p in eukaryotes in the regulation of their respective systems.
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
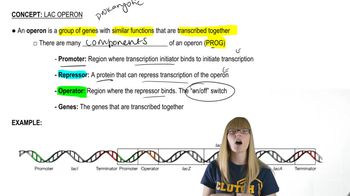
Lac Operon Induction in E. coli
Role of Gal3p in Yeast GAL System

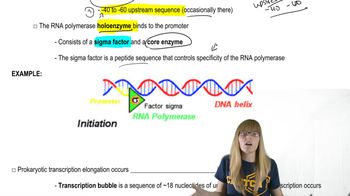
Differences Between Prokaryotic and Eukaryotic Transcriptional Regulation
A particular type of anemia in humans, called β-thalassemia, results from a severe reduction or absence of the normal β-globin chain of hemoglobin. However, the γ-globin chain, normally only expressed during fetal development, can functionally substitute for β-globin. A variety of studies have explored the use of the nucleoside 5-azacytidine for the expression of γ-globin in adult patients with β-thalassemia.
How might 5-azacytidine lead to expression of γ-globin in adult patients?
A particular type of anemia in humans, called β-thalassemia, results from a severe reduction or absence of the normal β-globin chain of hemoglobin. However, the γ-globin chain, normally only expressed during fetal development, can functionally substitute for β-globin. A variety of studies have explored the use of the nucleoside 5-azacytidine for the expression of γ-globin in adult patients with β-thalassemia.
Explain why this drug may also have some adverse side effects.
Regulation of the lac operon in E. coli and regulation of the GAL system in yeast are analogous in that they both serve to adapt cells to growth on different carbon sources. However, the transcriptional changes are accomplished very differently. Consider the conceptual similarities and differences as you address the following.
Compare and contrast the cis-regulatory elements of the lac operon and GAL gene system.
Regulation of the lac operon in E. coli and regulation of the GAL system in yeast are analogous in that they both serve to adapt cells to growth on different carbon sources. However, the transcriptional changes are accomplished very differently. Consider the conceptual similarities and differences as you address the following.
Compare and contrast how these two systems are negatively regulated such that they are downregulated in the presence of glucose.
DNA methylation is commonly associated with a reduction of transcription. The following data come from a study of the impact of the location and extent of DNA methylation on gene activity in eukaryotic cells. A bacterial gene, luciferase, was inserted into plasmids next to eukaryotic promoter fragments. CpG sequences, either within the promoter and coding sequence (transcription unit) or outside of the transcription unit, were methylated to various degrees, in vitro. The chimeric plasmids were then introduced into cultured cells, and luciferase activity was assayed. These data compare the degree of expression of luciferase with differences in the location of DNA methylation [Irvine et al. (2002). Mol. and Cell. Biol. 22:6689–6696]. What general conclusions can be drawn from these data?
