Early development depends on the temporal and spatial interplay between maternally supplied material and mRNA and the onset of zygotic gene expression. Maternally encoded mRNAs must be produced, positioned, and degraded [Surdej and Jacobs-Lorena (1998). Mol. Cell Biol. 18:2892–2900]. For example, transcription of the bicoid gene that determines anterior–posterior polarity in Drosophila is maternal. The mRNA is synthesized in the ovary by nurse cells and then transported to the oocyte, where it localizes to the anterior ends of oocytes. After egg deposition, bicoid mRNA is translated and unstable bicoid protein forms a decreasing concentration gradient from the anterior end of the embryo. At the start of gastrulation, bicoid mRNA has been degraded. Consider two models to explain the degradation of bicoid mRNA: (1) degradation may result from signals within the mRNA (intrinsic model), or (2) degradation may result from the mRNA's position within the egg (extrinsic model). Experimentally, how could one distinguish between these two models?

A number of genes that control expression of Hox genes in Drosophila have been identified. One of these homozygous mutants is extra sex combs, where some of the head and all of the thorax and abdominal segments develop as the last abdominal segment. In other words, all affected segments develop as posterior segments. What does this phenotype tell you about which set of Hox genes is controlled by the extra sex combs gene?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Hox Genes
Mutations and Phenotypes

Gene Regulation

Formation of germ cells in Drosophila and many other embryos is dependent on their position in the embryo and their exposure to localized cytoplasmic determinants. Nuclei exposed to cytoplasm in the posterior end of Drosophila eggs (the pole plasm) form cells that develop into germ cells under the direction of maternally derived components. R. Amikura et al. [(2001). Proc. Nat. Acad. Sci. (USA) 98:9133–9138] consistently found mitochondria-type ribosomes outside mitochondria in the germ plasma of Drosophila embryos and postulated that they are intimately related to germ-cell specification. If you were studying this phenomenon, what would you want to know about the activity of these ribosomes?
One of the most interesting aspects of early development is the remodeling of the cell cycle from rapid cell divisions, apparently lacking G1 and G2 phases, to slower cell cycles with measurable G1 and G2 phases and checkpoints. During this remodeling, maternal mRNAs that specify cyclins are deadenylated, and zygotic genes are activated to produce cyclins. Audic et al. [(2001). Mol. and Cell. Biol. 21:1662–1671] suggest that deadenylation requires transcription of zygotic genes. Present a diagram that captures the significant features of these findings.
The apterous gene in Drosophila encodes a protein required for wing patterning and growth. It is also known to function in nerve development, fertility, and viability. When human and mouse genes whose protein products closely resemble apterous were used to generate transgenic Drosophila [Rincon-Limas et al. (1999). Proc. Nat. Acad. Sci. (USA) 96:2165–2170], the apterous mutant phenotype was rescued. In addition, the whole-body expression patterns in the transgenic Drosophila were similar to normal apterous.
What is meant by the term rescued in this context?
The apterous gene in Drosophila encodes a protein required for wing patterning and growth. It is also known to function in nerve development, fertility, and viability. When human and mouse genes whose protein products closely resemble apterous were used to generate transgenic Drosophila [Rincon-Limas et al. (1999). Proc. Nat. Acad. Sci. (USA) 96:2165–2170], the apterous mutant phenotype was rescued. In addition, the whole-body expression patterns in the transgenic Drosophila were similar to normal apterous.
What do these results indicate about the molecular nature of development?
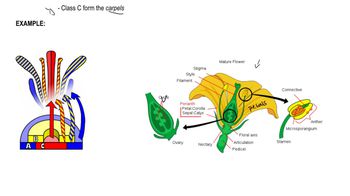
In Arabidopsis, flower development is controlled by sets of homeotic genes. How many classes of these genes are there, and what structures are formed by their individual and combined expression?
