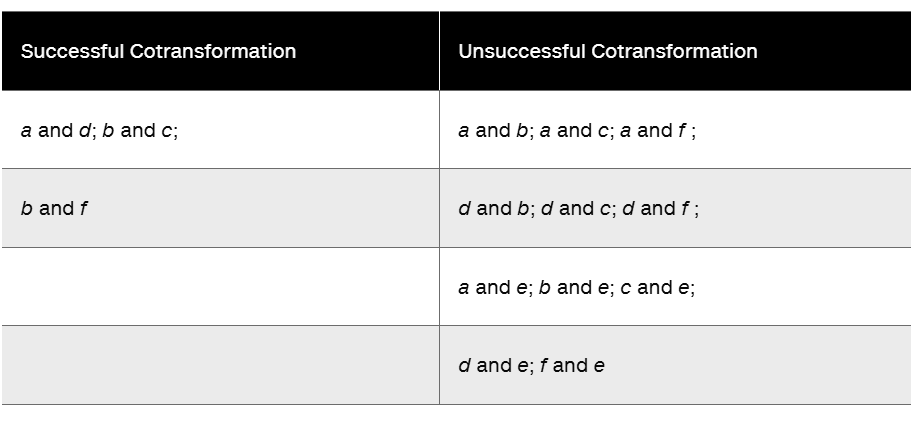
An Hfr strain is used to map three genes in an interrupted mating experiment. The cross is Hfr/a⁺b⁺c⁺ rif x F⁻/a⁻b⁻c⁻ rifT (No map order is implied in the listing of the alleles; rifT is resistance to the antibiotic rifampicin.) The a⁺ gene is required for the biosynthesis of nutrient A, the b⁺ gene for nutrient B, and the c⁺ gene for nutrient C. The minus alleles are auxotrophs for these nutrients. The cross is initiated at time = 0, and at various times, the mating mixture is plated on three types of medium. Each plate contains minimal medium (MM) plus rifampicin plus specific supplements that are indicated in the following table. (The results for each time interval are shown as the number of colonies growing on each plate.)
Based on these data, determine the approximate location on the chromosome of the a, b, and c genes relative to one another and to the F factor.